- *Corresponding Author:
- Zhong Zheng
Department of Dermatology, Gongan County People’s Hospital, Jinzhou, Hubei 434399, China
E-mail: 289096498@qq.com
| This article was originally published in a special issue, “Clinical Advancements in Life Sciences and Pharmaceutical Research” |
| Indian J Pharm Sci 2024:86(5) Spl Issue “42-53” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Drugs play an indispensable role in treating liver tumors nowadays. Meanwhile, liver tumors present a significant health challenge, demanding accurate diagnostic tools that are safe for all patients, including those with iodine allergies in pharmacy or renal insufficiency. Addressing the limitations of traditional contrast-enhanced computed tomography scans, we introduce plain scan liver tumors dataset and a new model based on the unit model (YNetr model), which is named for its resemblance to a Y rotating counterclockwise. The YNetr model is the plain scan liver tumors dataset consists of multiple liver tumor plain scan segmentation datasets, meticulously assembled and annotated. Our innovation, the YNetr model, leverages wavelet transforms to extract varied frequency information, aiming to enhance diagnostic accuracy without the need for contrast agents. This model achieved a remarkable dice coefficient of 62.63 % on the plain scan liver tumors dataset, outperforming existing models by 1.22 %. Our comprehensive comparison included models like UNet 3+XNet, UNetr, and more, highlighting YNetr’s superior capability in non-contrast liver tumor segmentation. This breakthrough not only provides a safer diagnostic alternative but also improves the effectiveness of drug treatments, demonstrating the vital role of technological innovations in improving patient treatment and safety.
Keywords
Plain scan liver tumors dataset, segmentation, artificial intelligence, dual-encoder, wavelet, computed tomography
Computed Tomography (CT) is a diagnostic technique that uses precisely collimated X-ray beams and highly sensitive detectors to perform sectional scans around a specific part of the human body. This method is characterized by its rapid scanning time and clear images. It can be applied to scan various parts of the body and holds immense clinical value for disease diagnosis. CT scans are increasingly used for abdominal diseases, primarily for diagnosing conditions related to the liver, gallbladder, pancreas, spleen, peritoneal cavity, retroperitoneal space, and the urinary and reproductive systems. They are particularly useful for diagnosing space-occupying lesions, inflammatory and traumatic changes.
Liver tumors refer to neoplasms occurring in the liver, which can be benign or malignant. Malignant liver tumors mainly include primary and secondary liver cancers, along with other malignancies like hepatoblastoma and sarcoma. Benign liver tumors include hemangioma, adenoma, and focal nodular hyperplasia.
Various diagnostic methods are available for liver tumors, such as ultrasound, CT, Magnetic Resonance Imaging (MRI), and even Positron Emission Tomography (PET)-CT. Currently, CT is the most widely used due to its ability to display liver crosssections every (0.5-1) cm, avoiding overlap from different angles of the liver. This technique reveals tumors and lesions within the liver, including their location, size, shape and relationship with surrounding tissues. CT enhancement is performed for further clarification when the nature of a lesion is difficult to determine.
In medical field, when a patient exhibits liver function abnormalities, employing CT scan to determine the type of liver disease is crucial. However, with the increasing workload on physicians, manually identifying liver tumors using visual inspection is extremely time-consuming. To save the diagnostic time, doctors can utilize artificial intelligence to segment lesions, aiding in diagnosis. Particularly in an era where deep learning is rapidly advancing, the effectiveness of artificial intelligence has been notably demonstrated. This has further propelled the application of artificial intelligence in medical imaging diagnostics.
In Contrast-Enhanced (CE) CT scans, iodine-based contrast agents are typically used, which can pose risks for certain patients, such as those with allergies to iodine or with renal insufficiency. On the other hand, plain CT scan, as a diagnostic method that does not require contrast agents and is a safer option for these patients. Here is more information about iodine-based contrast agents.
Iodine-based contrast agents, such as iodixanol are widely used in medical imaging, particularly in enhanced CT scans. They work by increasing the contrast between blood vessels and surrounding tissues, aiding doctors in observing and diagnosing conditions more clearly. Despite their critical role in medical diagnostics, iodine-based contrast agents can also bring certain risks and side effects.
A small percentage of patients may have an allergy to iodixanol or other iodine-based contrast agents, which could lead to rashes, urticaria, and in severe cases, anaphylactic shock.
For patients with pre-existing kidney issues, the use of iodine-based contrast can exacerbate renal strain, sometimes even leading to Acute Kidney Injury (AKI). Hence, kidney function is usually assessed before administering iodine-based contrast agents. Other potential side effects were also observed. Though less common, some patients might experience nausea, vomiting, headaches, or changes in taste after undergoing a CT scan with iodine-based contrast.
To mitigate these risks, physicians typically evaluate patient’s medical history for allergies and assess kidney function before proceeding with iodine-based contrast agents. In cases where the risk is deemed too high, alternative diagnostic methods, such as plain scan CT or MRI without contrast, may be considered to ensure patient safety.
However, in existing studies, all research on Liver Tumor Segmentation (LITS) is based on CECT LITS, with the LITS[1] dataset and related algorithms (such as nnU-Net[2]) being prominent examples. Despite this, research on plain scan CT, LITS remains limited, even though it holds clinical significance. Specifically, the clinical significance of plain scan CT, LITS are more as mentioned below.
Advantage of avoiding contrast agents CECT scans typically require iodine-based contrast agents, which can pose risks for certain patients (such as those with iodine allergies or renal insufficiency). Plain scan CT, as a diagnostic method that does not require contrast agents, is a safer choice for these patients.
Compared to enhanced scans, plain scan CT is generally less expensive and simpler to operate. In resource-limited areas (such as primary care hospitals) or in emergency situations, plain scan CT might be a more practical or faster option.
When the patients exhibit no obvious symptoms, plain scan CT can be used for early detection and monitoring of liver tumors.
Furthermore, developing algorithms capable of accurately identifying liver tumors from plain scan CT images demonstrates the progress of artificial intelligence and machine learning in the field of medical imaging. This could pave the way for future medical imaging analysis technologies.
From the perspective of existing semantic segmentation models, most models have only one branch as an encoder and one branch as a decoder[3]. In contrast, XNet uses two branches as encoders and two branches as decoders to capture features at different frequencies of the image and adjust outputs for semi-supervised segmentation. However, XNet still has its drawbacks, as mentioned below-
XNet uses two branches as decoders, but studies have shown that the outputs of XNet’s two branches are very similar. Therefore, it is necessary to merge the two decoder branches of XNet into one through feature fusion. Meanwhile, in semi-supervised tasks, using two decoders is justifiable because it allows for the comparison and adjustment of the outputs from both decoders to accommodate unlabeled data. However, in fully supervised tasks, employing two decoders is unreasonable. This is because the labels are already known, eliminating the need for additional adjustments.
XNet solely relies on Convolutional Neural Network (CNN) for feature extraction, which cannot capture long-term dependencies. To address this issue, considering the use of transformers as feature extractors is necessary.
To address these issues, we have developed the first plain scan LITS dataset, Plain Scan Liver Tumors (PSLT). Additionally, we introduced a model, YNetr, featuring two branches as encoders and one branch as a decoder. This model achieved a dice coefficient of 62.63 % (State-Of-The-Art (SOTA)) on our dataset. In summary, our contributions are as follows:
To address the gap in plain scan LITS datasets, we have developed the first dedicated dataset for this purpose, PSLT. It encompasses plain scan data from forty distinct patients, totaling 10 923 slices.
We propose a novel model, YNetr, which employs a dual-branch architecture as encoders for multi-level feature extraction and a singular branch as a decoder for feature fusion. Additionally, the vision transformer architecture, mirroring the UNETR structure[4,5], is utilized within the encoder to capture global features of images. The conclusive experimental results demonstrate that YNetr achieves SOTA performance on the PSLT dataset.
Materials and Methods
Proposed dataset PSLT:
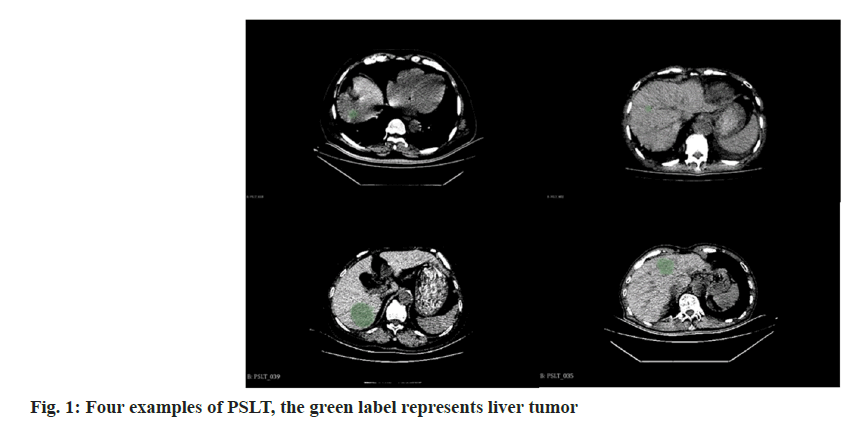
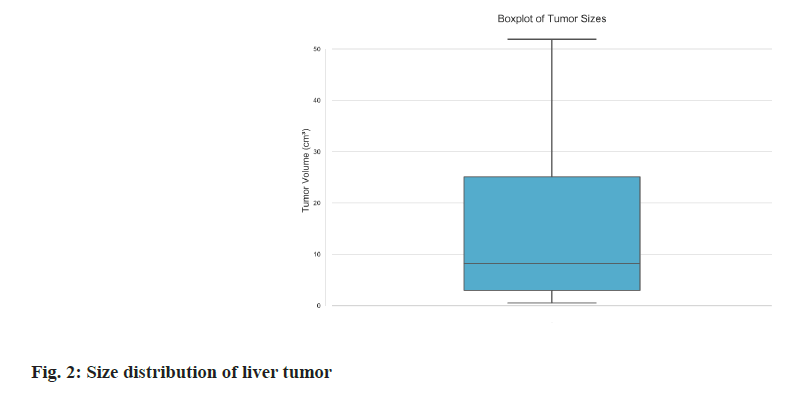
Dataset summary: The PSLT dataset consists of forty plain scan Three-Dimensional (3D) CT volumes collected from forty distinct patients by Gong’an County People’s Hospital, Hubei Province. This dataset includes a wide variety of cases such as abdominal scans, thoracoabdominal scans, tumors in different stages. Each volume was comprehensively scanned utilizing a SIEMENS CT scanner, ensuring consistent imaging quality. The volumes span an extensive range, encompassing 145 to 873 slices per volume, with each slice boasting a resolution of 512×512 pixels. This evidence the remarkable high-resolution nature of the PSLT dataset. While each volume uniformly includes liver imagery, the scanned regions exhibit considerable variation, including abdominal and thoracoabdominal scans as Table 1 and Table 2. This heterogeneity considerably enriches the dataset’s diversity, offering a robust basis for various analytical applications. Four illustrative examples from the PSLT dataset is showcased in fig. 1. To ensure patient confidentiality, all data were subjected to rigorous anonymization processes and received the necessary ethical committee approvals, thereby upholding stringent privacy standards. For research and development purposes, the PSLT dataset was partitioned into two subsets randomly: A training set consisting of 28 volumes (7667 slices) and a testing set comprising 12 volumes (3256 slices), as delineated in Table 3. We also show the size distribution of liver tumors in fig. 2. This indicates that half of the tumors range between 3 cm3 and 25 cm3, with a more frequent distribution of smaller tumors <8 cm3. The prevalence of these smaller tumors under 8 cubic centimeters adds complexity to the identification of lesions.
| Dataset | Plain scan | Liver tumor | Segmentation | Volume number | Modality |
|---|---|---|---|---|---|
| TCGA-LIHC | X | √ | X | 1688 | CT/MR |
| SILVER07 | X | X | √ | 30 | PT CT |
| LTSC’08 | X | √ | √ | 30 | CT |
| VISCERAL’16 | X | X | √ | 60/60 | CT/MRI |
| CHAOS19 | X | X | √ | 40/120 | CT/MRI |
| LiTS | X | √ | √ | 201 | CT |
| PSLT (Ours) | √ | √ | √ | 40 | CT |
Table 1: Comparison of Various Datasets for Liver Tumor Segmentation
| Abdominal | Thoracoabdominal | |
|---|---|---|
| Train set (n=28) | 20 | 8 |
| Test set (n=12) | 8 | 4 |
Table 2: Scan Site in PSLT
| Total slices | Maximum slices | Minimum slices | |
|---|---|---|---|
| Train set (n=28) | 7667 | 873 | 169 |
| Test set (n=12) | 3256 | 571 | 145 |
Table 3: Scan Site in PSLT
| Model | Dice coefficient |
|---|---|
| UNet 3+ | 58.43 |
| XNet | 54.56 |
| UNetr | 58.67 |
| Swin INetr | 58.89 |
| Trans BTS | 55.64 |
| COTr | 56.72 |
| nnUNetv2 (2D) | 44.31 |
| nnUNetv2 (3D fullers) | 60.21 |
| Med next (2D) | 46.98 |
| Med next (3D fullers) | 61.41 |
| YNetr (Our model) | 62.63 |
Table 4: Comparative Experiment Results, Bold Signifies SOTA Performance
Professional data annotation: Due to the complexity of 3D medical imaging data, manually annotating each frame of 3D medical images is extremely timeconsuming. To enhance the efficiency of annotation, we employed semi-automated techniques using the 3D Slicer software[6-10]. Specifically, a chief physician with >10 y of experience conducted the initial annotations using 3D Slicer.
Subsequently, these annotations were reviewed by a deputy chief physician with more than 20 y of experience. In cases of differing opinions, a consensus was reached through discussions involving multiple colleagues. The data labeling task will continue until the doctors believe there are no issues. During the annotation phase, each volume required 0.3 h-0.5 h for annotation by the chief physician and 0.05 h-0.2 h for review by the deputy chief physician, including discussions. Overall, approximately 3 mo were invested in the collection, annotation, and review of the PSLT dataset.
Compared to existing datasets, our dataset is the first to focus on non-contrast CT scans for LITS. A comparative analysis with other datasets is presented in Table 1, illustrating its unique position in the current research landscape.
Proposed model YNetr:
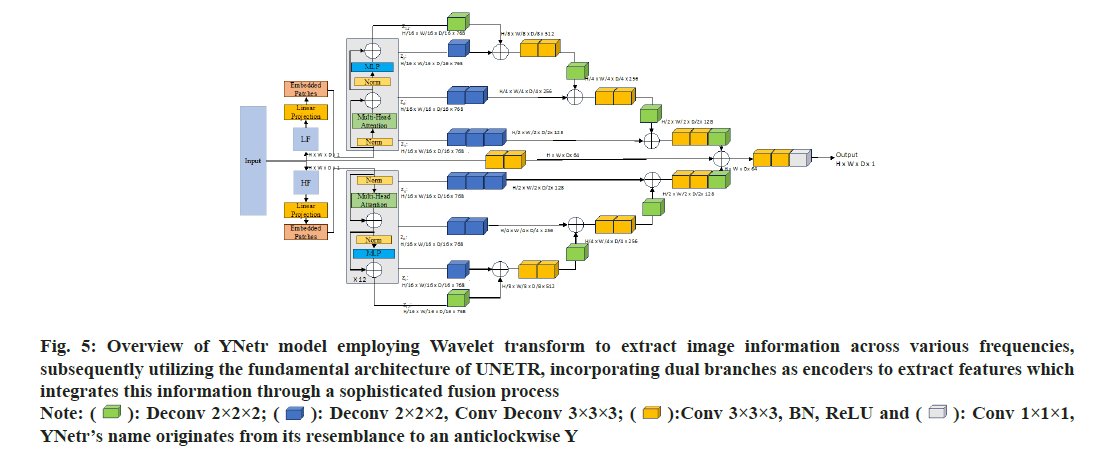
Framework overview: The YNetr architecture features an anticlockwise Y-shaped. It comprises two branches forming the encoder and a single branch as the decoder. Each branch utilizes the structure of UNETR. Within the encoder, a 1D sequence is generated from a 3D input volume(H×W×D×C), where (H,W,D) represents the height, width and depth, and C denotes the input channels. This sequence is formed by flattening uniformly non-overlapping patches x_v£R(N×(P3.C), where (P,P,P) indicates the dimensions of a patch and (N=(H×W×D)/P3) represents the length of the sequence. Different from the UNETR framework, the YNetr architecture innovatively utilizes a dual-branch encoder to capture medical imaging data across varied frequencies. In a distinctive approach to integrating encoder and decoder information, YNetr employs addition as its fusion technique, instead of the more conventional method of dimensional stacking. This design choice facilitates a more seamless and effective integration of multi-scale features by capturing different frequency information, enhancing the model’s capacity to process complex medical images.
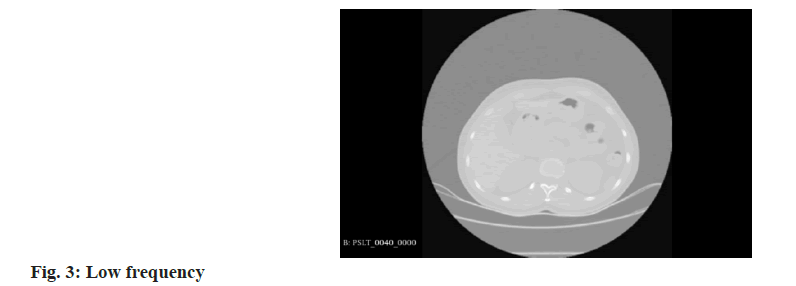
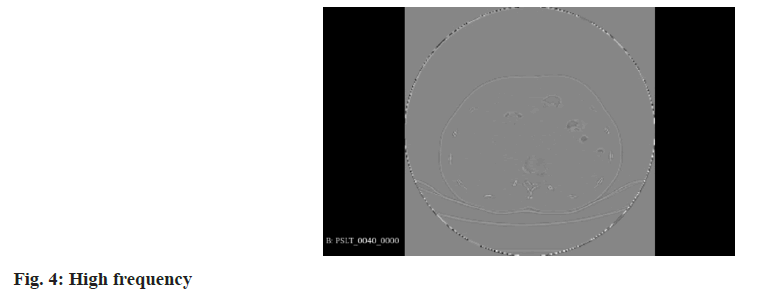
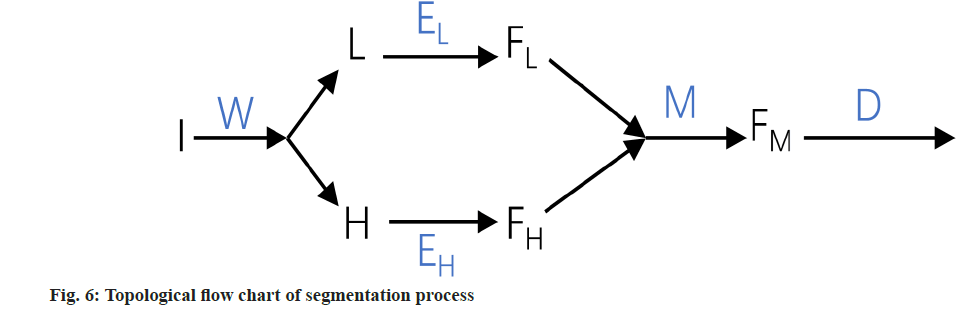
Wavelet transform: In the realm of 3D medical imaging, data fundamentally represents discrete signals encompassing information across various frequencies. The wavelet transform is adept at segregating this multi-frequency information effectively. This transformation is applied to partition raw image data into distinct components, namely Low Frequency (LF), and High Frequency (HF) in three orientations: Horizontal, vertical, and diagonal. These are technically denoted as R (raw image) for LF. H, V, and D are denoted for horizontal, vertical, and diagonal HF components, respectively. These components capture the low-frequency signals along with high-frequency information in different orientations. For the comprehensive representation of high-frequency data, it is essential to amalgamate these directional high-frequency components. The formulation of low and high-frequency information is delineated as follows:
LF=R
HF=H+V+D
Where, low-frequency information is characterized by reduced noise and fewer details, enhancing the clarity of the overarching structure. In contrast, the high-frequency information provides more noise but clearer object boundaries, as depicted in the accompanying figure as shown in fig. 3 and fig. 4.
Details of YNetr: To capture the global information of an image, each branch of the encoder incorporates twelve layers of the UNetr block. Specifically, at the 3rd, 6th, 9th, and 12th layers, outputs are generated with dimensions of H/16×W/16×D/16×768 by unfolding.
In other layers of the encoder, the output will be taken as the input for the next layer in the form of N×(P3.C) where (N=(H×W×D)/P3), (P=16) and (C=768). Post convolution, these dimensions are transformed to H/8×W/8×D/8×512, H/8×W/8×D/8×512, H/4×W/4×D/4×256, and H/2×W/2×D/2×128 respectively. During the decoder phase (the central line in fig. 5), convolutional operations are applied for up sampling to restore the image to its original size. Additionally, given that the model incorporates two encoders but only a single decoder, we opt for additive fusion rather than dimensional stacking for skip connections, thereby integrating the encoder information into the decoder; fig. 6 presents the topological flow chart of YNetr.
Fig 5: Overview of YNetr model employing Wavelet transform to extract image information across various frequencies, subsequently utilizing the fundamental architecture of UNETR, incorporating dual branches as encoders to extract features which integrates this information through a sophisticated fusion process  YNetr’s name originates from its resemblance to an anticlockwise Y
YNetr’s name originates from its resemblance to an anticlockwise Y
Loss function:
In our experiments, the loss function is crucial for guiding the segmentation task towards optimal performance. We have employed a combined dice and Cross-Entropy (CE) loss function. This hybrid loss function leverages the advantages of both the dice loss, which is proficient in handling class imbalance by measuring overlap, and the CE loss, which robustly penalizes incorrect predictions on a voxel-wise basis. The formulation of our adopted loss function is as follows:
LDice-CE (G,Y)=αLDice (G,Y)+(1-α) LCE (G,Y)
Where, (G) represents the ground truth and (Y) denotes the predicted segmentation. The parameter Alpha (α) strikes a balance between the two loss components. In this case, we define (α) as 1/2 by experiments. Specifically, the Dice loss (LDice) is defined by:
LDice (G,Y)=1-(2Σi Gi Yi )/( Σi Gi+Σi Yi)
And CE loss (LCE) is given by,
LCE (G,Y)=-Σi Gi log (Yi)
Here, (i) indices over all voxels, (Gi) denotes the ground truth value and (Yi) represents the predicted probability for each voxel.
Evaluation metrics:
The evaluation of segmentation models is pivotal to our study. To quantitatively assess the performance of our proposed model, we employ the Dice coefficient, a widely recognized metric for segmentation tasks. The Dice coefficient, also known as the Dice similarity index, measures the overlap between the predicted segmentation and the ground truth. It is particularly effective for medical image segmentation where binary classification predominates. The Dice coefficient is mathematically defined as:
D (G, Y)=2×TP/2×TP+FP+FN
Where, (G) stands for the ground truth binary mask, (Y) signifies the predicted segmentation mask, True Positives (TP), False Positives (FP) and False Negatives (FN).
Implementation details:
The implementation of our YNetr model was conducted utilizing the PyTorch framework alongside MONAI, a medical open network for AI, which provided a robust and flexible platform for our deep learning architecture. We used four NVIDIA GeForce RTX 3090 graphics cards for training. The model optimization was carried out using the AdamW optimizer, with the initial learning rate set at 0.0001 and the training carried out for over 300 epochs.
Within the encoder module, the patch resolution was calibrated to a 16×16×16 matrix. During the inference stage, a sliding window approach was employed, and the overlap rate was methodically set to 0.5 to ensure comprehensive coverage and accuracy. Here, the data were segmented into slices of 128×128×128 and subsequently fed into the model. To circumvent the model’s propensity to learn excessively from the background, we maintained a balanced ratio of positive to negative samples at 1:1. Additionally, to mitigate central bias, random translations of 48×48×48 blocks were implemented.
Our conclusive experiments revealed that our model achieved a Dice coefficient precision of 62.63 %, surpassing the performance of models in other comparative studies.
Results and Discussion
In our comparative experiments in Table 4, several models were deployed to assess their performance: UNet 3+[11], XNet[3], UNetr[5], Swin UNetr[12], TransBTS[12], COTr[13], nnUNetv2 (2D), nnUNetv2 (3D Fullers)[2], MedNext (2D), and MedNext (3D Fullers)[14]. Among these, Mednext (3D Fullers) had superior performance, achieving a Dice score of 61.41 %. However, our model excelled by attaining a Dice coefficient accuracy of 62.63 %, thereby surpassing the existing model MedNext (3D Fullers) which stood at 61.41 %. The data from the comparative experiments are delineated in the table below.
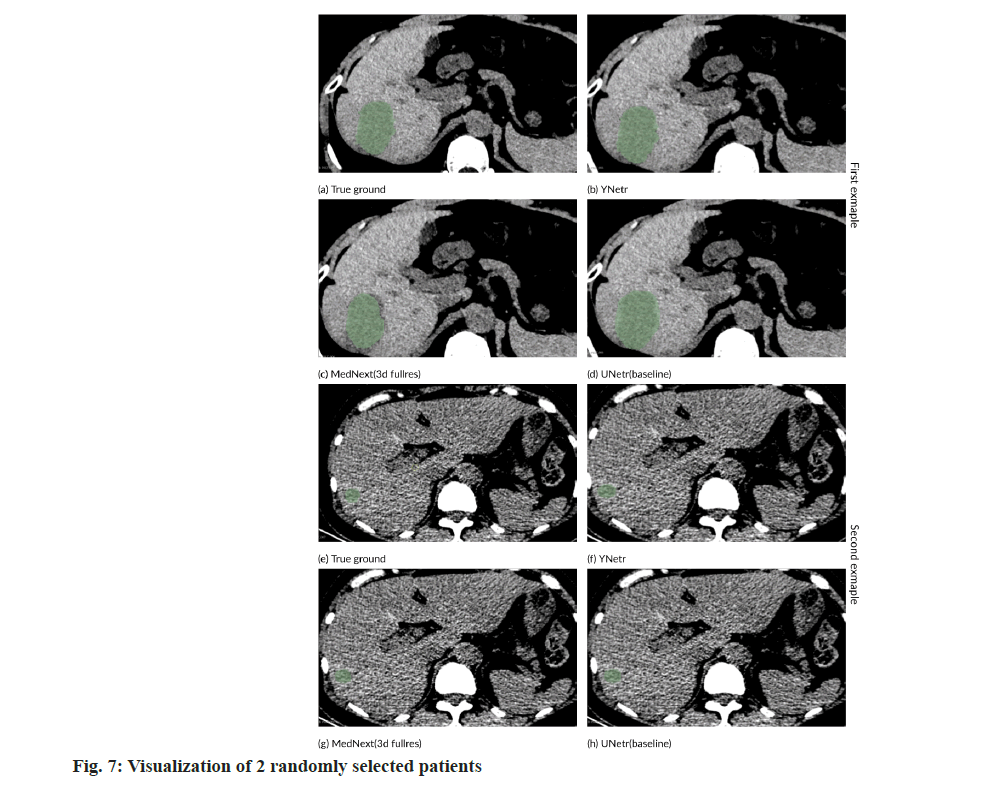
In fig. 7, we present a comparative segmentation illustration on the PSLT dataset between MedNext and YNetr. The visual comparison clearly demonstrates the superior segmentation efficacy of YNetr over MedNext, as evidenced by the enhanced delineation of the segmented regions.
In our research, we empirically established a correlation between the patch size and the accuracy of the segmentation results. Specifically, our experiments demonstrate that as the patch size increases, there is a corresponding decrease in segmentation accuracy which is shown in fig. 5. For instance, with a patch size set to 16×16×16, our model achieved a Dice coefficient of 62.63 %. In contrast, increasing the patch size to 32×32×32 resulted in a lower Dice coefficient of 61.08 %.
To validate the efficacy of the transformer module in feature extraction, we conducted a series of comparative experiments. We experimented with substituting both branches of the dual-encoder architecture entirely with CNN architectures. This configuration yielded an accuracy of 57.35 %. By contrast, utilizing the transformer for extracting lowfrequency features and CNNs for high-frequency information, we achieved an accuracy of 60.97 %. Conversely, employing CNNs for low-frequency and transformers for high-frequency feature extraction resulted in an accuracy of 59.04 %. Both configurations fell short of the 62.63 % accuracy attained when transformers were used exclusively in both branches. These results underscore the superiority of transformers in extracting both low and high-frequency information. The ablation study results are visually presented in Table 5 for an intuitive comparison.
| Modules | Dice coefficient (%) |
|---|---|
| Two encoders with CNN | 57.35 |
| LF (CNN) and HF (16 patches transformer) | 59.04 |
| LF (16 patches transformer) and HF (CNN) | 60.97 |
| Two encoders with 32 patches transformer | 61.08 |
| Two encoders with 16 patches transformer | 62.63 |
Table 5: Ablation Experiments
| Loss function | Dice coefficient(%) |
|---|---|
| Dice loss | 62.54 |
| CE | 62.36 |
| Boundary loss | 62.24 |
| Dice-CE loss | 62.63 |
Table 6: Segmentation Accuracy of Different Loss Functions
In the ablation studies, we evaluated the impact of different loss functions on the performance of our proposed model as shown in Table 6. We considered several commonly used loss functions in medical image segmentation, including Dice Loss, CE Loss, a combination of Dice and CE Loss (Dice-CE), and boundary loss. The effectiveness of each loss function was measured based on the segmentation accuracy, quantified by the Dice coefficient percentage.
Notably, the Dice-CE Loss outperformed the other loss functions, achieving the highest accuracy of 62.63 %. This superior performance can be attributed to the balanced combination of Dice and CE Loss. The Dice component of the loss function effectively handles the issue of class imbalance prevalent in medical image datasets, particularly in cases where the region of interest occupies a small portion of the image. Meanwhile, the CE component contributes to robust voxel-wise error penalization, ensuring that each pixel’s classification is accurately accounted for. The synergistic effect of combining these two loss functions leads to improved segmentation performance, as evidenced by our experimental results. This underscores the efficacy of the Dice- CE loss in our task, providing an optimal balance between class imbalance handling and precise voxelwise classification.
While extensive research has been conducted on datasets for enhanced CT LITS, there is a notable gap in the literature regarding CT non-contrast datasets for this purpose. Existing field is the LTSC’08 segmentation challenge organized by The Cancer Imaging Archive (TCIA)[7], which released 30 enhanced CT voxel datasets specifically for LITS. The LITS dataset from the Technical University of Munich (TUM), consisting of 201 voxel datasets[1], stands as a benchmark for LITS in enhanced CT imaging.
Complementing these, a variety of datasets from different institutions contribute to the breadth of research in this domain. For example, the TCGA-LIHC dataset from TCIA provides a substantial volume of data with 1688 instances[6], although it does not include segmentation labels. The DKFZ institution’s SILVER07 dataset presents an additional 30 CT volumes[7]. Siemens and the University of Geneva, with their respective datasets, contribute further to the field, though their focus is not exclusively on LITS. 60 scans with two modalities (MRI and CT) for segmentation and landmark detection in anatomical structure were provided by the VISCERAL taset includes CHAOS with provides 40 CT volumes and 120 MRI volumes[9]. The comparison of these datasets with the PSLT dataset is shown in Table 1.
Since the introduction of the UNet architecture[15], a multitude of semantic segmentation methods based on UNet have been developed, particularly for 3D voxel data. VNet[16], which utilizes 3D CNNs for feature extraction, marked a significant advancement, heralding a new phase in semantic segmentation methods for 3D voxel data. The advent of the vision transformer led to its adoption in various methodologies[4], such as TransU-Net[17], nn-former[18], CoTr[14], TransBTS[13], Transfuse[19], and UNETR each achieving commendable results[5]. Following the swin transformer’s emergence, sliding-window techniques have been implemented in the medical image segmentation field, with swin UNETR showing impressive efficacy across multiple datasets[20]. Later, to make the model more lightweight, Slim UNETR has been proposed[12]. The introduction of nnUNet[2] and mednext[14] provided a significant boon for those less proficient in AI. These systems, through high-level integration and data augmentation, have demonstrated strong performance in numerous tasks. In 2023, XNet was proposed, a novel approach that extracts features from datasets at varying frequencies and employs two branches each for the encoder and decoder. This methodology has achieved SOTA results in a wide range of semi-supervised and fully supervised semantic segmentation tasks.
Incorporating the wavelet transform, renowned for its exceptional frequency and spatial analysis capabilities, into Deep Neural Networks (DNNs) has seen various explorations for semantic segmentation tasks, as evidenced in research works[21-26]. The primary approaches involve leveraging the wavelet transform for either pre-processing or postprocessing tasks[21,22], as well as substituting specific CNN layers (notably those responsible for upsampling and down-sampling) with wavelet-based operations[23-25]. Despite these advancements, the applicability of these methods tends to be confined to particular types of segmentation targets, thereby constraining their widespread utility.
A study introduced a symmetric CNN architecture augmented with wavelet transform[27], named aerial LaneNet, aimed at enhancing lane-marking semantic segmentation in aerial images. Additionally, the concept of wavelet constrained pooling layers, as an alternative to traditional pooling mechanisms for the segmentation of synthetic aperture radar imagery, was presented in CWNN[28]. Furthermore, Wave SNet employs wavelet transforms for the meticulous extraction of image nuances during the downsampling phase and utilizes the inverse wavelet transform to restore these details in the up-sampling process[29].
The advantages of plain CT over CECT are primarily evident in several key aspects, plain scan CT are more convenient because they do not require the injection of contrast media, thus avoiding the associated complications such as contrast media extravasation, allergy and nephropathy. In contrast, CECT requires contrast injection. Iodinated Contrast Media (ICM) is one of the most frequently administered[30], AKI is a potential complication of intravascular iodinated contrast exposure, which usually presents as a transient small decrease in renal function that occurs within a few days of contrast administration and is associated with serious adverse outcomes, including progressive renal dysfunction and death[31]. It occurs in >30 % of patients after intravenous ICM and causes serious complications[32]. Intravenous administration of a contrast agent is required to assess blood flow to the lesion, and this may cause harm.
Plain CT scanning is a time-efficient, single-step procedure, and rapid imaging equipment can complete the process in seconds, making it more acceptable to non-compliant patients. This minimizes the patient’s exposure to ionizing radiation. For example, while minimizing radiation exposure, Ultra-Low Dose (ULD) CT could facilitate the clinical implementation of large-scale lung cancer screening[33]. However, CECT tends to be lengthier than other imaging techniques due to multiple scanning phases, including non-contrast, arterial, venous, and sometimes delayed phases. This poses greater challenges for non-compliant patients, increases exposure time, and results in higher radiation doses, which can lead to greater potential harm.
Cost considerations indicate that plain scan CT are less expensive than CECT. This is because plain CT only involves the fee for the CT procedure itself, whereas CECT involves the fee for the enhanced scan and the contrast agent.
Plain scan CT are more suitable for screening during health check-ups, as they are generally more acceptable to routine patients than CECT. Wholebody CT enables the identification of a significant number of relevant and early findings, which increase significantly with age, leading to changes in lifestyle and early treatment[34].
YNetr model, with its Dual-Encoder architecture for PSLT, marks a pivotal advancement in medical imaging, especially from a pharmacological perspective. By bypassing the need for iodinebased contrast agents, it addresses significant patient safety concerns, particularly for those with allergies or renal insufficiency. This innovation not only enhances patient care by reducing the risks associated with contrast media but also aligns with the principles of pharmacoeconomic by potentially lowering healthcare costs through the avoidance of adverse reactions and the associated care expenses. Furthermore, YNetr’s approach is instrumental in the push towards personalized medicine, allowing for safer, more accurate diagnostics across a broader patient demographic. Its development underscores the critical role of interdisciplinary collaboration, merging insights from pharmacology, radiology, and computational science to improve diagnostic methodologies. As such, YNetr’s contribution to non-contrast imaging demonstrates a significant stride towards safer, more efficient, and patientcentric diagnostic solutions, reflecting the growing intersection between pharmacological safety and technological innovation in healthcare.
Plain scan CT cannot accurately assess vascular anomalies such as aneurysms, embolisms, or aortic dissections without contrast media. Lesions may not be discernibly contrasted against surrounding normal tissue, which hinders the ability to display the lesion’s structure and internal composition. This limitation can lead to imprecise assessments of lesion size, location, and type, increasing the risk of missed diagnoses or misdiagnoses. Low contrast resolution can make it challenging to differentiate between various structures, such as necrotic or cystic changes, and to detect small pathologies like lesions. In addition, plain scan CT do not allow observation of vascular features, contrast uptake patterns and relationships with surrounding structures, which are necessary to characterize some lesions and therefore CECT is often required for accurate diagnosis. This leads to less comprehensive information, which hinders the conclusive diagnosis and differentiation of benign or malignant lesions. This is not only a diagnostic challenge for the physician, but also a challenge for the artificial intelligence in the segmentation of the lesions in the plain scans. Besides, large-scale clinical and imaging modalities, particularly radiological features of CECT, can be integrated to predict the clinical prognosis of patients with Microvascular Invasion (MVI) and Hepatocellular Carcinoma (HCC)[35]. For this reason, we recommend that further research be conducted into the cost-effective segmentation of liver tumors on plain scans.
In conclusion, while identifying liver tumors in plain scans may be more challenging than in CECT imaging, the benefits of lower duration, cost, and reduced harm position the segmentation of liver tumors in plain scans as a promising research area. Preoperative CT features can be used to characterize the macro trabecular-massive subtype and the vessels that encapsulate tumor clusters pattern. These features have prognostic significance for early recurrence in patients with HCC[36].
In the segmentation of liver tumors, there is a significant density difference between plain and enhanced CT scans. Compared to enhanced CT, plain CT scans are more challenging to discern. To overcome this difficulty, using wavelet transform to capture varying density information is a good approach. However, determining the most suitable density information and methods for information fusion still requires further research. From a pharmacological perspective, this approach also mitigates the risks associated with the use of contrast agents, which can be particularly relevant for patients with allergies or renal insufficiency, emphasizing the importance of developing safer imaging alternatives. Additionally, since our PSLT dataset contains only 40 volumes of liver tumor annotations, expanding the dataset and annotating other abdominal regions also necessitates further study. This expansion is crucial not only for enhancing the dataset’s comprehensiveness but also for exploring the potential of non-contrast imaging techniques in a wider range of pharmacological and medical applications.
In conclusion, this paper presents the YNetr model, which employs a dual-transformer architecture as the encoder, tested on the first non-contrast LITS dataset PSLT, achieving SOTA results. In summary, our work not only introduces the inaugural non-contrast LITS dataset for medical research, providing a benchmark accuracy for subsequent studies but also adopts a dual-encoder approach to fuse information across different frequencies, offering researchers novel insights into feature extraction methodologies. This contribution is particularly significant in the context of pharmacology, where the need for non-invasive, safe, and effective diagnostic methods is everpresent. By addressing the challenges of plain CT imaging and emphasizing the reduction of contrast agent use, the YNetr model stands as a promising step toward safer and more accurate diagnostic practices in medical and pharmacological research.
Acknowledgements:
This retrospective study was approved by the Biological and Medical Ethics Committee of Gong’an County People’s Hospital.
Author’s contributions:
Wen Sheng, Jun Zhao and Zhengdi Sima have contributed equally to this work and they are considered as first authors. Zhong Zheng and Zhihong Zhang contribute the same to the article and are the corresponding authors.
Conflict of interests:
The authors declared no conflict of interests.
References
- Bilic P, Christ P, Li HB, Vorontsov E, Ben-Cohen A, Kaissis G, et al. The liver tumor segmentation benchmark (lits). Med Image Anal 2023;84:102680.
- Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat Method 2021;18(2):203-11.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Huang J, Wang C, Song L, Yang G. Xnet: Wavelet-based low and high frequency fusion networks for fully-and semi-supervised semantic segmentation of biomedical images. Proc IEEE/CVF Int Conference Comput Vision 2023:21085-96.
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv Preprint arXiv 2010.11929. 2020.
- Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, et al. Unetr: Transformers for 3d medical image segmentation. Proc IEEE/CVF winter conference on applications of computer vision 2022 (pp. 574-584).
- Erickson B, Kirk S, Lee Y, Bathe O, Kearns M, Gerdes C, et al. Radiology data from the cancer genome atlas liver hepatocellular carcinoma collection. Cancer Imag Arch 2016;10:K9.
- Heimann T, Van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans Med Imag 2009;28(8):1251-65.
[Crossref] [Google Scholar] [PubMed]
- Jimenez-del-Toro O, Müller H, Krenn M, Gruenberg K, Taha AA, Winterstein M, et al. Cloud-based evaluation of anatomical structure segmentation and landmark detection algorithms: VISCERAL anatomy benchmarks. IEEE transactions on medical imaging 2016;35(11):2459-75.
[Crossref] [Google Scholar] [PubMed]
- Kavur AE, Gezer NS, Bar?? M, Aslan S, Conze PH, Groza V, et al. CHAOS challenge-combined (CT-MR) healthy abdominal organ segmentation. Med Image Anal 2021;69:101950.
[Crossref] [Google Scholar] [PubMed]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 2012;30(9):1323-41.
[Crossref] [Google Scholar] [PubMed]
- Huang H, Lin L, Tong R, Hu H, Zhang Q, Iwamoto Y, et al. Unet 3+: A full-scale connected unet for medical image segmentation. ICASSP 2020:1055-9.
- Hatamizadeh A, Nath V, Tang Y, Yang D, Roth HR, Xu D. Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. Int MICCAI Brainlesion Workshop 2021:272-84.
- Xie Y, Zhang J, Shen C, Xia Y. Cotr: Efficiently bridging cnn and transformer for 3D medical image segmentation. MICCAI
- Roy S, Koehler G, Ulrich C, Baumgartner M, Petersen J, Isensee F, et al. Mednext: transformer-driven scaling of convnets for medical image segmentation. Int Conference Med Image Comput Assisted Interv 2023:405-15.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. MICCAI 2015:234-41.
- Milletari F, Navab N, Ahmadi SA. V-net: Fully convolutional neural networks for volumetric medical image segmentation. 2016 Fourth Int Conference 2016: 565-71.
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv: 2021;2102:04306.
- Zhou HY, Guo J, Zhang Y, Yu L, Wang L, Yu Y. Nnformer: Interleaved transformer for volumetric segmentation. arXiv Preprint arXiv 2021;2109:03201.
- Zhang Y, Liu H, Hu Q. Transfuse: Fusing transformers and cnns for medical image segmentation. MICCAI 2021:14-24.
- Pang Y, Liang J, Huang T, Chen H, Li Y, Li D, et al. Slim UNETR: Scale hybrid transformers to efficient 3d medical image segmentation under limited computational resources. IEEE Trans Med Imaging 2023;43(3):994-1005.
- Yin X, Xu X. A method for improving accuracy of deeplabv3+ semantic segmentation model based on wavelet transform. Int Conference Commun Signal Proc Syst 2021:315-20.
- Upadhyay K, Agrawal M, Vashist P. Wavelet based fine-to-coarse retinal blood vessel extraction using U-net model. 2020 Int Conference SPCOM 2020:1-5.
- Zhao C, Xia B, Chen W, Guo L, Du J, Wang T, Lei B. Multi-scale wavelet network algorithm for pediatric echocardiographic segmentation via hierarchical feature guided fusion. Appl Soft Computing 2021;107:107386.
- Sinha P, Wu Y, Psaromiligkos I, Zilic Z. Lumen & media segmentation of ivus images via ellipse fitting using a wavelet-decomposed subband cnn. 2020 IEEE 30th Int Workshop MLSP 2020:1-6.
- Gao F, Wang X, Gao Y, Dong J, Wang S. Sea ice change detection in SAR images based on convolutional-wavelet neural networks. IEEE Geosci Remote Sensing Lett 2019;16(8):1240-4.
- Liu P, Zhang H, Lian W, Zuo W. Multi-level wavelet convolutional neural networks. IEEE Access 2019;7:74973-85.
- Azimi SM, Fischer P, Körner M, Reinartz P. Aerial LaneNet: Lane-marking semantic segmentation in aerial imagery using wavelet-enhanced cost-sensitive symmetric fully convolutional neural networks. IEEE Transactions Geosci Remote Sensing 2018;57(5):2920-38.
- Duan Y, Liu F, Jiao L, Zhao P, Zhang L. SAR image segmentation based on convolutional-wavelet neural network and Markov random field. Pattern Recog 2017;64:255-67.
- Li Q, Shen L. Wavesnet: Wavelet integrated deep networks for image segmentation. Chin Conference PRCV 2022;17:325-37.
- Macdonald DB, Hurrell CD, Costa AF, McInnes MD, O’Malley M, Barrett BJ, et al. Canadian association of radiologists guidance on contrast-associated acute kidney injury. Can J Kidney Health Dis 2022;9:499-514.
[Crossref] [Google Scholar] [PubMed]
- Cashion W, Weisbord SD. Radiographic contrast media and the kidney. Clin J Am Soc Nephrol 2022;17(8):1234-42.
[Crossref] [Google Scholar] [PubMed]
- Lin Q, Li S, Jiang N, Shao X, Zhang M, Jin H, et al. PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation. Redox Biol 2019;26:101254.
[Crossref] [Google Scholar] [PubMed]
- Jiang B, Li N, Shi X, Zhang S, Li J, de Bock GH, et al. Deep learning reconstruction shows better lung nodule detection for ultra-low-dose chest CT. Radiology 2022;303(1):202-12.
[Crossref] [Google Scholar] [PubMed]
- Millor M, Bartolome P, Pons MJ, Bastarrika G, Beloqui O, Cano D, et al. Whole-body computed tomography: A new point of view in a hospital check-up unit? Our experience in 6516 patients. La Radiol Med 2019;124:1199-211.
[Crossref] [Google Scholar] [PubMed]
- Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol 2019;70(6):1133-44.
[Crossref] [Google Scholar] [PubMed]
- Feng Z, Li H, Zhao H, Jiang Y, Liu Q, Chen Q, et al. Preoperative CT for characterization of aggressive macrotrabecular-massive subtype and vessels that encapsulate tumor clusters pattern in hepatocellular carcinoma. Radiology 2021;300(1):219-29.
[Crossref] [Google Scholar] [PubMed]