- *Corresponding Author:
- Tong Li
The Third Central Clinical College of Tianjin Medical University, China
E-mail: xingyu7892024@163.com
| Date of Received | 25 February 2023 |
| Date of Revision | 17 November 2023 |
| Date of Acceptance | 03 May 2024 |
| Indian J Pharm Sci 2024;86(3):839-850 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Although new cell-based research suggests a link between PPFIA4 and colon adenocarcinoma cancer, there is currently no pan-cancer study available. Using The Cancer Genome Atlas datasets, we firstly investigated the possible oncogenic function of PPFIA4 in 33 tumors. We discovered that PPFIA4 is substantially expressed in most malignancies and is associated with tumor patient prognosis. Furthermore, missense mutation and amplification of the PPFIA4 gene were the most common types of genetic change. In contrast to lung squamous cell carcinoma, tumor purity in prostate adenocarcinoma was strongly negatively connected with PPFIA4 expression and considerably positively correlated with the amount of infiltration of cluster of differentiation 8+ T cells, dendritic cells, macrophages, cluster of differentiation 4+ T cells, B cells and neutrophils in the tumor microenvironment. PPFIA4 expression in bladder cancer and lung squamous cell carcinoma is a strong correlation with the expression levels of tumor microenvironment, M1 and M2 macrophages, which are seen in the majority of monocyte marker groups. Furthermore, the functional mechanisms of PPFIA4 were influenced by chemical synaptic transmission as well as the development of the nervous system. In summary, our work provided essential insights into PPFIA4 dysregulation in pan-cancer and prompted potential molecular mechanisms in the progression of cancer, which highlight its potential therapeutic target for patients.
Keywords
PPFIA4, pan-cancer, immune microenvironment, prognosis, The Cancer Genome Atlas
Since tumorigenesis is a complex process with multiple factors including genes, therefore, studying the mechanism of genes is very important to evaluating tumor and clinical prognosis. The publicly funded The Cancer Genome Atlas (TCGA) project has provided us with important help for pan-cancer analysis, including the relevant functional genomics data set of 33 tumors[1-4].
Human PPFIA4 (AF034801) is a 5'-truncated part of complementary Deoxyribonucleic Acid (cDNA). Exons 1-29 of PPFIA4 are found in the human genome sequences AL451082.6 and AC096632.3. The PPFIA4-MYOG locus and the PPFIA2-LIN7a-MYF5-MYF6 locus are para- homologous. The composition of PPFIA family members is composed of three Sterile Alpha Motif (SAM) and PFIH1-4 domains. The VRTYSC motif (the C-terminal binding domain of Glutamate Receptor-Interacting Protein (GRIP)) is contained in PPFIA1-A3, but not in PPFIA4. The binucleate location signal is contained in the PFIH4 domain[5].
Compared with PPFIA1-3, there are relatively few studies on PPFIA4, but there are still many important studies showing that PPFIA4 is involved in the process of cancer glycolysis and also shows that it is an important gene[6-8]. However, based on relevant clinical data, there is no evidence of a pan-carcinogenic association between PPFIA4 and various tumor types. Our study is the first to use the TCGA project for pan-carcinomatous analysis of PPFIA4. We conducted the first pan-cancer analysis of PPFIA4 using survival status, immune infiltration, gene expression, gene changes and associated cellular pathways in the TCGA project. To explore the relationship between different cancer pathogenesis and PPFIA4, provide potential molecular mechanisms for clinical prognosis and immunotherapy.
Materials and Methods
PPFIA4 expression analysis:
First, we use the Tumor Immune Evaluation Resource (TIMER) 2.0 website for data retrieval and enter PPFIA4 in the "Gene DE" module. The differences in the expression of PPFIA4 in various related tumors and normal tissues were detected by using TIMER software. The Gene Expression Profiling Interactive Analysis 2 (GEPIA2) website provides further information on tumors that lack normal tissue[9]. We used the GEPIA database to observe the expression of PPFIA4 in various cancers at different clinic pathological stages.
Survival prognosis analysis:
GEPIA2's "Survival Map" module is needed to retrieve the Overall Survival (OS) and Disease- Free Survival (DFS) of various cancers in the TCGA database. Using the GEPIA2 information, we were able to create a survival map.
Genetic alteration analysis:
Genetic changes in PPFIA4 were obtained using cBioPortal[10,11]. Inquire about genetic variation characteristics by selecting "TCGA Pan-Cancer Atlas Studies" and "PPFIA4". The "Cancer type Summary" module was used to obtain the results of copy number alteration, mutation type and change frequency for different tumors in all TCGA databases. The "mutations" module allows you to acquire a schematic representation of the protein structure including the mutation site of PPFIA4, which is shown below. We used the "compare" module to collect information on survival differences in relevant tumor cases in the TCGA database.
Immune infiltration analysis:
After screening out fibroblasts associated with immunotumor, TIMER2 was used to analyze the relationship between tumor immune invasions. In the aspect of immune penetration, we adopted MCPCOUNTER, EPIC, XCELL and Tracking of Indels by Decomposition (TIDE) algorithms for research. The data of immune infiltration analysis is displayed by heat map and scatter plot.
TIMER database analysis:
TIMER is different cancer type immune system analysis infiltration of comprehensive resources (https://cistrome.shinyapps.io/timer/)[12]. In terms of tumor-infiltrating immune cells, gene expression data should be analyzed in combination with relevant studies and the statistical method of TIMER[13]. We investigated the expression of PPFIA4 in TCGA samples from 33 different cancer types, as well as the relationship between the quantity of immune infiltration, macrophages. We used TIMER to conduct our investigation. The degree of gene expression in tumor purity is shown in the left-most panel of the fig. 1[14].
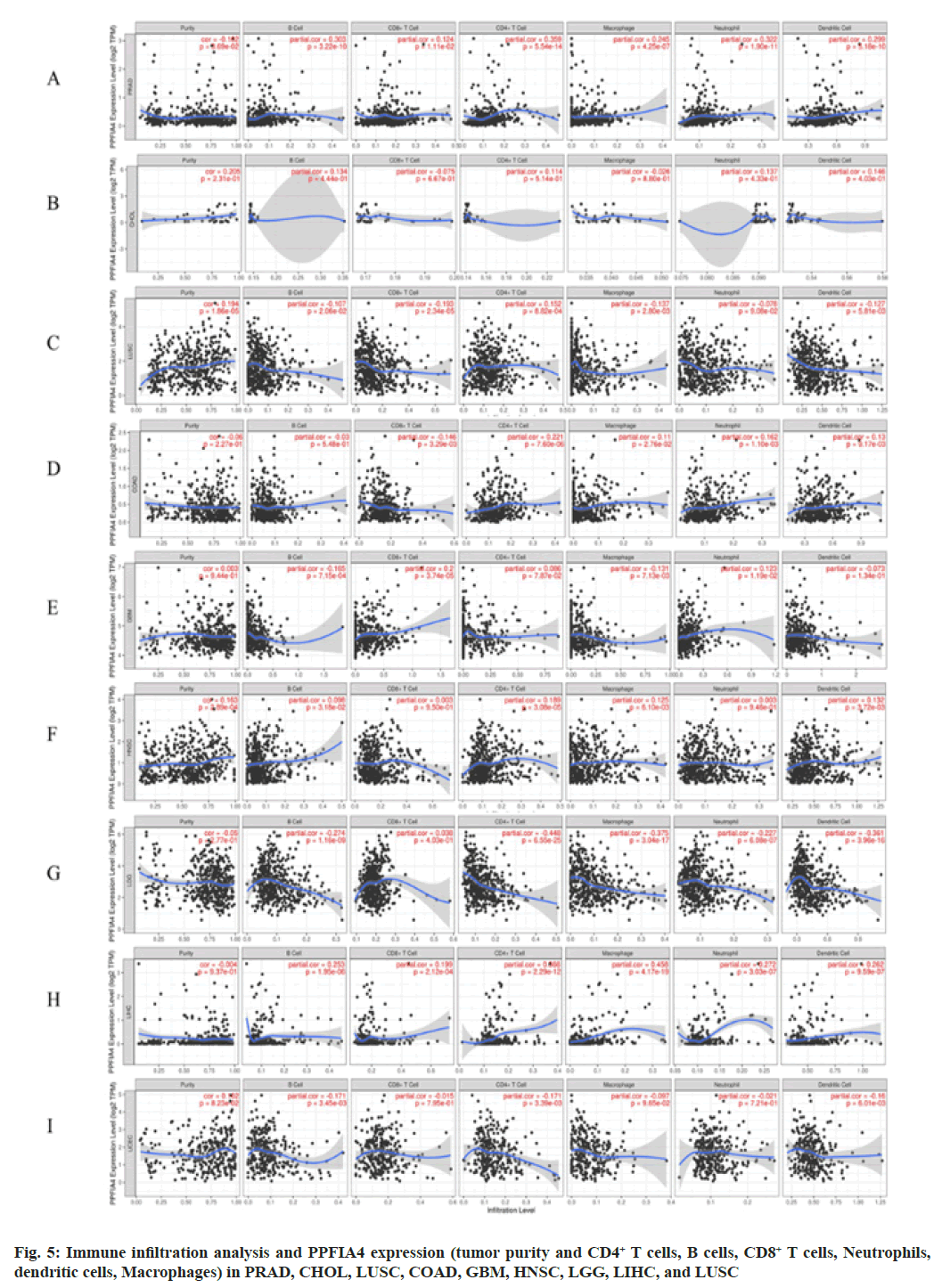
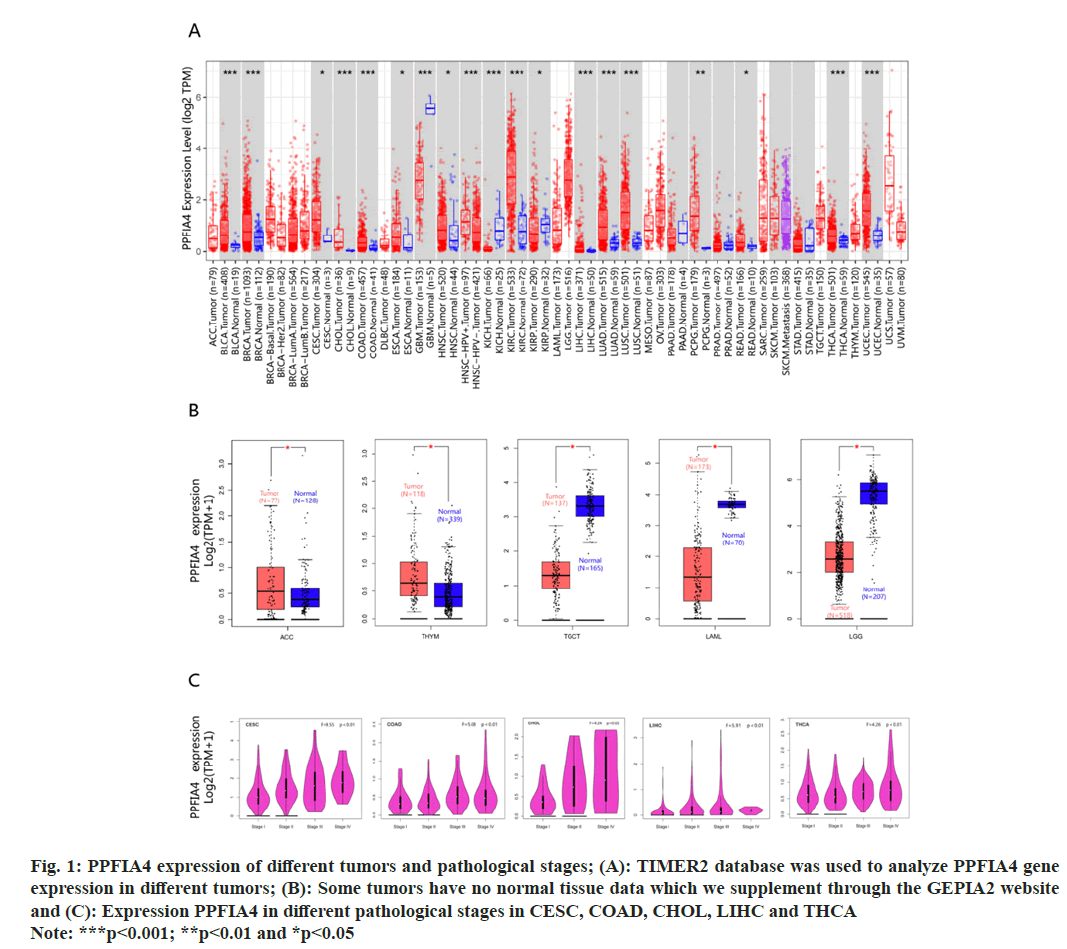
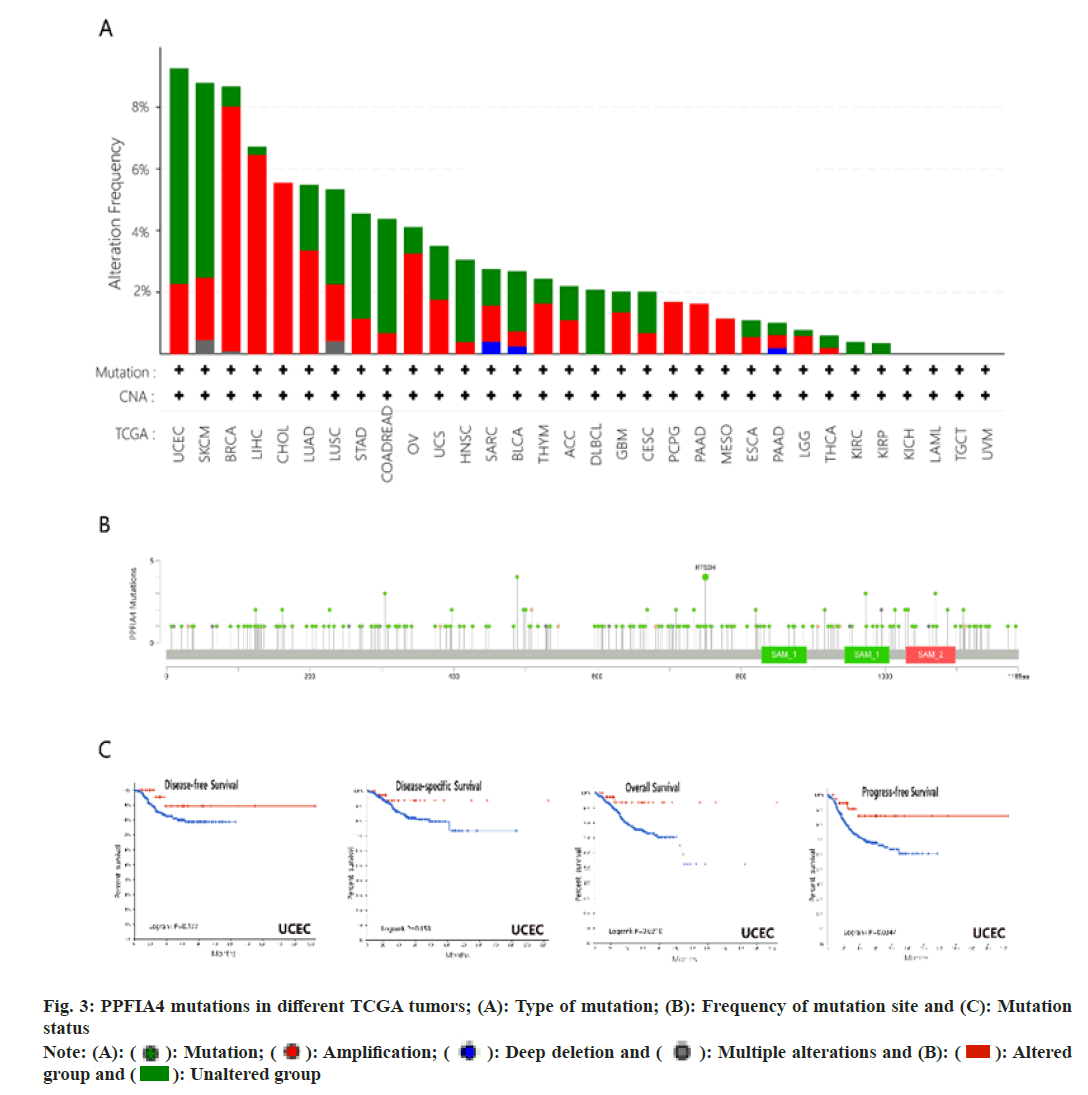
Fig. 1: PPFIA4 expression of different tumors and pathological stages; (A): TIMER2 database was used to analyze PPFIA4 gene
expression in different tumors; (B): Some tumors have no normal tissue data which we supplement through the GEPIA2 website
and (C): Expression PPFIA4 in different pathological stages in CESC, COAD, CHOL, LIHC and THCA
Note: ***p<0.001; **p<0.01 and *p<0.05
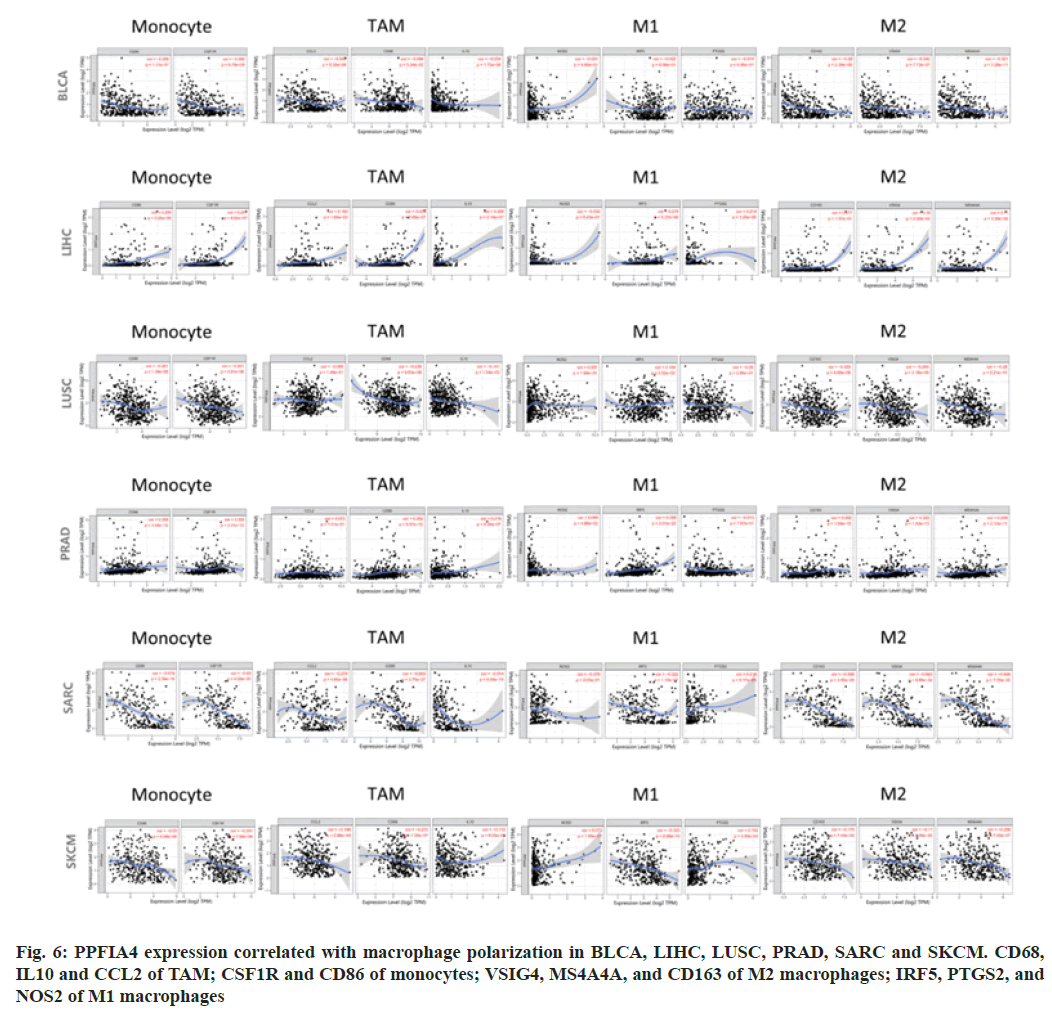
Studies have shown tumor-infiltrating immune cell markers[15-18]. Monocytes include CD86 and Colony Stimulating Factor Receptor (CSF-1R); M1 macrophages include Prostaglandin-Endoperoxide Synthase (PTGS2), Nitric Oxide Synthase 2 (NOS2) and Interferon Regulatory Factor 5 (IRF5); Tumor Microenvironment (TME) includes Interleukin 10 (IL-10), Clusters of Differentiation (CD) 68 and C-C Motif Chemokine Ligand 2 (CCL2); M2 macrophages include V-Set Ig-Domain-Containing 4 (VSIG4), CD163 and MS4A4A. The scatter plot is generated using Spearman correlation in a given tumor type.
Pathway analysis:
For protein interactions, log in to the STRING database to set parameters (https://string-db.org/). Select the evidence for the first item meaning of network edges, select the experiment for the second item of active interaction sources, and select the third parameter as 0.150. Finally, the interactor parameter is 50. TCGA's top 100 targeted genes in tumors and normal tissues related to PPFIA4 are obtained through the "Similar Gene Detection" module. The Database for Annotation, Visualization and Integrated Discovery (DAVID) database is used for the enrichment analysis of related pathways for the first 100 PPFIA4 targeted genes that have been identified.
Results and Discussion
We used the TIMER database to display the PPFIA4 expression status of TCGA tumor tissue in fig. 1A. The Genotype-Tissue Expression (GTEx) data set is used to supplement normal tissues for the expression level of PPFIA4. Testicular Germ Cell Tumor (TGCT), Acute Myeloid Leukemia (LAML), Thymoma (THYM), Brain Low-Grade Glioma (LGG), and Adrenocortical Carcinoma (ACC) tumor tissues have differences in PPFIA4 expression from normal tissues (fig. 1B, p<0.05). However, there are still some tumors that do not express significantly different, such as ovarian serous cystadenocarcinoma (OV), Sarcoma (SARC), Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (DLBC), Uterine Carcinosarcoma (UCS), or Breast Invasive Carcinoma (BRCA).
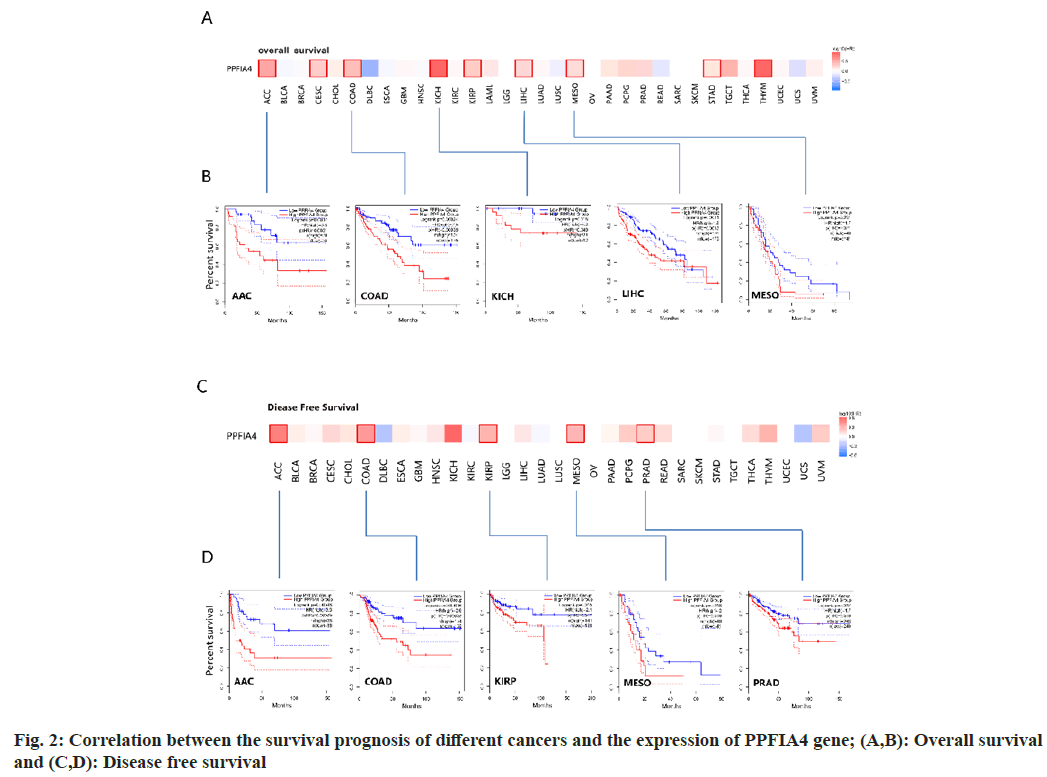
The "pathological stage map" module was used. We found that the pathological stages of Liver Hepatocellular Carcinoma (LIHC), Colon Adenocarcinoma (COAD), Cervical Squamous Cell Carcinoma (CESC), Thyroid Carcinoma (THCA), and Cholangiocarcinoma (CHOL) are closely related to the expression of PPFIA4 (fig. 1C, all p<0.05), but the correlation with other tumors is poor. We found that the OS rate of highly expressed PPFIA4 is poor, such as Augmentative and Alternative Communication (AAC), COAD, Kidney Chromophobe (KICH), LIHC, MESO, CESC, Kidney Renal Papillary Cell Carcinoma (KIRP), STAD and THYM (fig. 2A and fig. 2B, and all p<0.05).
In terms of DFS, the results showed that tumors with high PPFIA4 expression are closely related to poor prognoses, such as AAC, COAD, KIRP, MESO and Prostate Adenocarcinoma (PRAD) (fig. 2C and fig. 2D). According to the prognostic findings, the expression of PPFIA4 on DFS and OS was consistent, suggesting that the gene is reasonably stable and has a high reference value. There is no statistical difference between the expression of PPFIA4 in other tumors and DFS or OS.
Fig. 3A shows that the main types of "mutation" are Uterine Corpus Endometrial Earcinoma (UCEC) and SKCM, and PPFIA4 has the highest change frequency (>8 %). The main types of "amplification" are BRCA (>6 %) and LIHC. Interestingly, the only type of change in CHOL, Pheochromocytoma and Paraganglioma (PCPG), PAAD and MESO cases is “amplification”. Fig. 3B shows the sites and type of genetic changes in PPFIA4. We found that the genetic changes of PPFIA4 are mainly missense mutations and amplifications. The frameshift mutation of the PPFIA4 gene is induced by the R750H change in the Tudor domain (fig. 3B). In CUEC, patients with PPFIA4 mutations have a better prognosis in terms of Progression-Free Survival (PFS) and OS. However, the prognosis for Disease-Specific Survival (DSS) and DFS was same when compared to those with no PPFIA4 alterations (fig. 3C).
In the TME, tumor infiltrating immune cells participate in many processes such as tumor genesis, metastasis and development, which is the key link of tumor cell proliferation[19-22]. Tumor-associated fibroblasts control the behavior of different tumor-infiltrating immune cells[23-26]. We use the EPIC, XCELL, MCPCOUNTER and TIDE algorithms among others (fig. 4A). After further analysis, based on our findings, we concluded that the estimated infiltration value of cancer-related fibroblasts into COAD, Esophageal Carcinoma (ESCA), Human Papillomavirus (HPV) (-), Head and Neck Squamous Cell Carcinoma (HNSC), OV, and STAD, TCGA tumors were statistically significantly linked with the expression of PPFIA4 (fig. 4B).
We further investigated whether the level of immune infiltration of 33 cancer types is related to the expression of PPFIA4. According to the results of the aforementioned investigation, the amount of CD4+ T cell infiltration in 20 malignancies is substantially linked with the expression of the transcription factor PPFIA4. In addition, the infiltration level of B cells (15 cancers), tumor purity (14 cancers), CD8+ T cell infiltration (11 cancers), macrophages (15 cancers), dendritic cells (12 cancers), and neutrophils (14 cancers) was shown to be substantially linked with the expression of PPFIA4 (fig. 5).
Interestingly, our study discovered that tumor purity was considerably negatively connected with PPFIA4 expression and was significantly favorably correlated with the infiltration level of neutrophils, CD8+ T cells, macrophages, B cells, dendritic cells, and CD4+ T cells in PRAD (fig. 5). However, the amount of immune cell infiltration or tumor purity was not substantially linked with the expression of PPFIA4 in CHOL (fig. 5). Furthermore, although the degree of neutrophil infiltration in Lung Squamous Cell Carcinoma (LUSC) was not substantially connected with PPFIA4 expression, it was strongly inversely correlated with B cells, macrophages, CD8+ T cells, and dendritic cells (fig. 5).
We looked at the association between various immune infiltrating cells and PPFIA4 expression to see whether there was a link between the two, including CD68, IL10, and CCL2 of Tumor- Associated Macrophages (TAMs); PTGS2, IRF5, and NOS2 of M1; CSF1R and CD86 of monocytes; VSIG4, MS4A4A and CD163 of M2 (fig. 6).
Our study showed that most of the marker sets were expressed at levels of monocytes, M2 macrophages, M1 macrophages and TAMs were strongly correlated with the expression of PPFIA4 in Bladder Cancer (BLCA), LIHC, LUSC, PRAD, SARC, SKCM, COAD, HNSC, LGG, SKCM-M, THYM and UCUE (p<0.05; fig. 6).
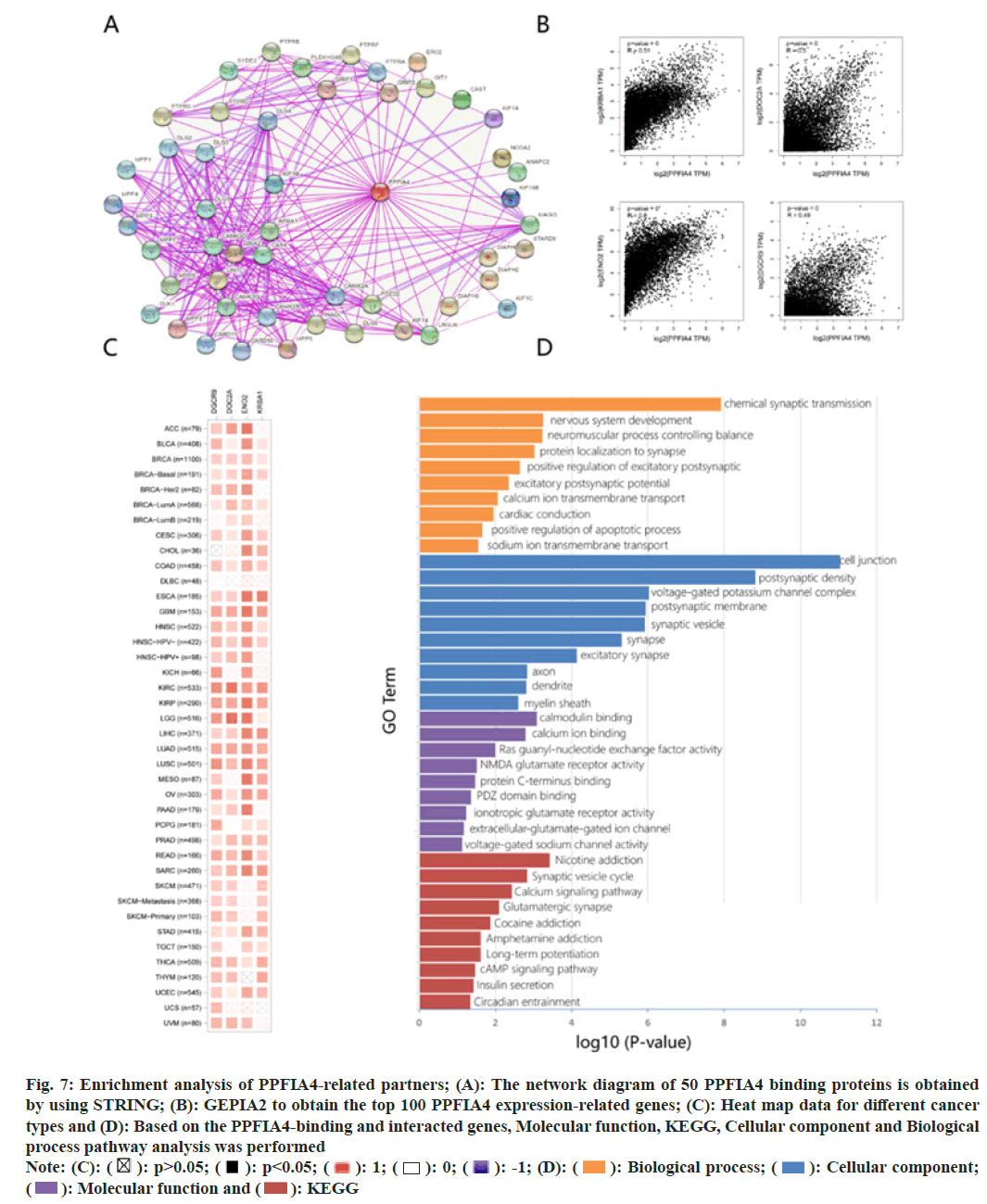
We conduct pathway enrichment analysis by screening targeted PPFIA4 expression-related genes and PPFIA4 binding proteins. To further analyze the relationship between the molecular mechanism of tumorigenesis and PPFIA4. The network diagram of 50 PPFIA4 binding proteins is obtained by using STRING (fig. 7A).
Fig. 7: Enrichment analysis of PPFIA4-related partners; (A): The network diagram of 50 PPFIA4 binding proteins is obtained
by using STRING; (B): GEPIA2 to obtain the top 100 PPFIA4 expression-related genes; (C): Heat map data for different cancer
types and (D): Based on the PPFIA4-binding and interacted genes, Molecular function, KEGG, Cellular component and Biological
process pathway analysis was performed
Note: (C):  Biological process;
Biological process;  Cellular component;
Cellular component;  KEGG
KEGG
After obtaining the top 100 PPFIA4 expression- related genes, we utilized GEPIA2 to further refine our search. On the right is a representation of the top four most highly expressed genes (fig. 7B); the expression levels of KRBA1, DOC2A, NEO2 and DGCR9 are positively associated with the expression of PPFIA4 in the vast majority of cancers. The heat map data shows a positive correlation (fig. 7C).
The molecular function data of fig. 7D suggest that “calmodulin binding, calcium ion binding” and “glutamate” may have a role in PPFIA4's influence on molecular function. The KEGG data in fig. 7D shows that the calcium signaling pathway and synaptic vesicle cycle suggest the involvement of PPFIA4 on cell junctions. Cell composition data show that PPFIA4 is related to chemical synaptic transmission or cell connection, such as chemical synaptic transmission, synaptic vesicle, synapse, postsynaptic membrane, postsynaptic density, excitatory synapse, voltage-gated potassium channel complex and others. Fig. 7D shows that the effect of PPFIA4 on tumor pathogenesis may be related to chemical synaptic transmission and nervous system development.
The PPFIA family has different physiological functions in humans, including PPFIA1, PPFIA2, PPFIA3, and PPFIA4[5]. Studies have shown that by up-regulating PPFIA1 in breast cancer, it is found that PPFIA1 as a promoter is involved in cancer cell invasion and migration[27]. In terms of cannabis-induced neuropsychological function, PPFIA2 expression was closely related to Cannabidiol (CBD)[28]. Detecting the expression of PPFIA3 methylation in serum samples, it provides a basis for a non-invasive gastric cancer detection program[29]. PPFIA4 is expressed in human skeletal muscle, heart and brain to varying degrees[5]. There are very little researche on PPFIA4 at present[30]. We have searched PPFIA4 on PubMed and have got 18 articles in search, only 3 of them are about cancer and PPFIA4[31]. For publications on PPFIA4 and pan-cancer analysis, we cannot obtain search information from a tumor perspective. There is no research showing an association between pan-cancer and PPFIA4. Whether PPFIA4 acts through some common molecular mechanism in the pathogenesis of different tumors remains to be answered. There is currently no research showing an association between pan-cancer and PPFIA4. Therefore, using TCGA database resources, the correlation analysis of PPFIA4 genes in 33 different tumors was carried out, including gene expression, genetic changes, related pathways, immune infiltration, and survival status.
Using the TIMER2 method, we were able to determine the expression status of PPFIA4 in TCGA samples of different cancer types. In 33 tumors, there was a significant difference in the expression of PPFIA4 in normal and tumor tissues and the expression of PPFIA4 in most tumors was higher than that in normal tissues (23 tumors) (fig. 1A), which may indicate that the expression of PPFIA4 in tumors has important significance. It lays a solid foundation for our follow-up research. Interestingly, we found that in AAC, COAD, KICH, LIHC, the expression of PPFIA4 in different stages of tumors is different, and in terms of OS rate, the low expression of PPFIA4 and the high expression of PPFIA4 in these four tumors There are also differences in PPFIA4 (p<0.05). Preliminary explanations are related to the expression of PPFIA4 in terms of tumor staging and OS. Although the expression of PPFIA4 in stage IV of AAC and stage III of KICH is lower than in the other three stages, it may be the reason for the small amount of data. In terms of OS, the greater the level of PPFIA4 expression, the worse the prognosis is considered to be.
We also have an important aspect of research on the relationship between immune invasion in cancer and PPFIA4, especially in COAD, PRAD, Glioblastoma Multiforme (GBM), LGG, UCEC, HNSC, LIHC and LUSC. Our results showed that the expression level of PPFIA4 was positively correlated with the infiltration levels of B cells, dendritic cells, CD8+ T cells, neutrophils, macrophages and CD4+ T cells in HNSC, LIHC and PRAD. Dendritic cells, neutrophils, macrophages, and B cells infiltrate at varying degrees in LGG, LUSC and UCEC, and their levels of infiltration are moderate to strongly negatively correlate with the amount of PPFIA4 expression. Furthermore, the link between immune cell marker genes and PPFIA4 expression shows that PPFIA4 is a critical regulator of SARC and LGG immunology, according to the findings. In terms of correlation study of immunological indicators, PPFIA4 expression was shown to have a poor association with M1 macrophage gene markers such as IRF5 and PTGS2, according to the results. In the M2 macrophage markers, MS4A4A and VSIG4 showed moderate to strong correlation (fig. 6). These results imply that PPFIA4 may have a regulatory function in the polarization of TAMs. Following Huang's findings, methylation levels and PPFIA4 expression in colon cancer tissues at various stages were shown to be greater than those found in normal tissues, and the presence of a high PPFIA4 expression level was found to be positively connected with poor patient survival. It is consistent with the findings of our investigation. PPFIA4 expression was shown to be positively correlated with Enolase 2 (ENO2) and PFKFB3 levels. Inhibition of ENO2 and PFKFB3 expression can reduce cell proliferation, invasion, glycolysis and migration, suggesting that PPFIA4 may be a potential biomarker for colon cancer[8].
Through the enrichment analysis in this study, we were able to determine the possible effect of chemical synaptic transmission, calcium signaling pathway, and nervous system development in tumor related etiology or important pathogenesis. A recent study showed that pathway enrichment analysis identified four biological processes and 20 pathways that were statistically significant, including neuron projection morphogenesis and modulation of excitatory postsynaptic potential[32]. These data are consistent with our analysis that PPFIA is closely related to neurite triggering. Although it studies the relationship between autism and genetic variation, our data analysis believes that it will be further confirmed by future studies on tumor and PPFIA4 expression.
In summary, we firstly performed the comprehensive pan-cancer investigation of PPFIA4 and discovered that the expression of PPFIA4 is statistically linked with genes expressed and changes, survival status, cell pathways and immune infiltration in various cancers. This information is useful in understanding the function of PPFIA4 expression in tumor development when seen through the lens of clinical tumor specimens, presenting a potential prognosis biomarker and immunotherapy target.
Acknowledgements:
This work was sponsored by Tianjin Science and Technology Project (21JCYBJC01590) and Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-035A).
Author’s contributions:
Yu Xing and Zili Zhang wrote the paper, collated and analyzed the data; Weiliang Song and Wenqing Gao guide paper writing and data analysis, Tong li design research plan, guide the completion of the paper.
Conflict of interests:
The authors declared no conflict of interests.
References
- Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell 2018;173(2):530.
- Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol 2015;19:A68-77.
- Wang Z, Jensen MA, Zenklusen JC. A practical guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol 2016;1418:111-41.
[Crossref] [Google Scholar] [PubMed]
- Zhang Z, Li H, Jiang S, Li R, Li W, Chen H, et al. A survey and evaluation of Web-based tools/databases for variant analysis of TCGA data. Brief Bioinform 2019;20(4):1524-41.
[Crossref] [Google Scholar] [PubMed]
- Katoh M, Katoh M. Identification and characterization of human PPFIA4 gene in silico. Int J Mol Med 2003;12(6):1009-14.
[Google Scholar] [PubMed]
- Xu F, Xu H, Li Z, Huang Y, Huang X, Li Y, et al. Glycolysis-based genes are potential biomarkers in thyroid cancer. Front Oncol 2021;11:534838.
[Crossref] [Google Scholar] [PubMed]
- Xu Z, Xu L, Liu L, Li H, Jin J, Peng M, et al. A glycolysis-related five-gene signature predicts biochemical recurrence-free survival in patients with prostate adenocarcinoma. Front Oncol 2021;11:625452.
[Crossref] [Google Scholar] [PubMed]
- Huang J, Yang M, Liu Z, Li X, Wang J, Fu N, et al. PPFIA4 promotes colon cancer cell proliferation and migration by enhancing tumor glycolysis. Front Oncol 2021;11:653200.
[Crossref] [Google Scholar] [PubMed]
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucl Acids Res 2019;47(W1):W556-60.
[Crossref] [Google Scholar] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signaling 2013;6(269):pl1.
[Crossref] [Google Scholar] [PubMed]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401-4.
[Crossref] [Google Scholar] [PubMed]
- Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77(21):e108-10.
[Crossref] [Google Scholar] [PubMed]
- Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol 2016;17(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun 2015;6(1):8971.
[Crossref] [Google Scholar] [PubMed]
- Siemers NO, Holloway JL, Chang H, Chasalow SD, Ross-MacDonald PB, Voliva CF, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PloS One 2017;12(7):e0179726.
[Crossref] [Google Scholar] [PubMed]
- Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer 2017;5:18.
[Crossref] [Google Scholar] [PubMed]
- Sousa S, Maatta J. The role of tumour-associated macrophages in bone metastasis. J Bone Oncol 2016;5(3):135-8.
- Tran Janco JM, Lamichhane P, Karyampudi L, Knutson KL. Tumor-infiltrating dendritic cells in cancer pathogenesis. J Immunol 2015;194(7):2985-91.
[Crossref] [Google Scholar] [PubMed]
- Steven A, Seliger B. The role of immune escape and immune cell infiltration in breast cancer. Breast Care 2018;13(1):16-21.
[Crossref] [Google Scholar] [PubMed]
- Garaud S, Buisseret L, Solinas C, Gu-Trantien C, de Wind A, van den Eynden G, et al. Tumor-infiltrating B cells signal functional humoral immune responses in breast cancer. JCI Insight 2019;4(18):e129641.
[Crossref] [Google Scholar] [PubMed]
- Baba Y, Nomoto D, Okadome K, Ishimoto T, Iwatsuki M, Miyamoto Y, et al. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci 2020;111(9):3132-41.
[Crossref] [Google Scholar] [PubMed]
- Deng X, Lin D, Zhang X, Shen X, Yang Z, Yang L, et al. Profiles of immune-related genes and immune cell infiltration in the tumor microenvironment of diffuse lower-grade gliomas. J Cell Physiol 2020;235(10):7321-31.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Song E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18(2):99-115.
[Crossref] [Google Scholar] [PubMed]
- Wang SS, Liu W, Ly D, Xu H, Qu L, Zhang L. Tumor-infiltrating B cells: Their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol 2019;16(1):6-18.
[Crossref] [Google Scholar] [PubMed]
- Kato T, Noma K, Ohara T, Kashima H, Katsura Y, Sato H, et al. Cancer-associated fibroblasts affect intratumoral CD8+ and FoxP3+ T cells via IL6 in the tumor microenvironment. Clin Cancer Res 2018;24(19):4820-33.
[Crossref] [Google Scholar] [PubMed]
- Suzuki J, Aokage K, Neri S, Sakai T, Hashimoto H, Su Y, et al. Relationship between podoplanin-expressing cancer-associated fibroblasts and the immune microenvironment of early lung squamous cell carcinoma. Lung Cancer 2021;153:1-10.
[Crossref] [Google Scholar] [PubMed]
- Dancau AM, Wuth L, Waschow M, Holst F, Krohn A, Choschzick M, et al. PPFIA1 and CCND1 are frequently coamplified in breast cancer. Genes Chromosomes Cancer 2010;49(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- He Y, de Witte LD, Schubart CD, Van Gastel WA, Koeleman BP, de Jong S, et al. Liprin alfa 2 gene expression is increased by cannabis use and associated with neuropsychological function. Eur Neuropsychopharmacol 2019;29(5):643-52.
[Crossref] [Google Scholar] [PubMed]
- Li WH, Zhou ZJ, Huang TH, Guo K, Chen W, Wang Y, et al. Detection of OSR2, VAV3, and PPFIA3 methylation in the serum of patients with gastric cancer. Dis Marker 2016;2016:5780538.
[Crossref] [Google Scholar] [PubMed]
- Fiorini N, Canese K, Starchenko G, Kireev E, Kim W, Miller V, et al. Best match: New relevance search for pubmed. PLoS Biol 2018;16(8):e2005343.
[Crossref] [Google Scholar] [PubMed]
- Low SK, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genetics 2017;49(6):953-8.
[Crossref] [Google Scholar] [PubMed]
- Jiao J, Zhang M, Yang P, Huang Y, Hu X, Cai J, et al. Analysis of common genetic variants associated with neuro-synapse development among 60 family trios affected with sporadic autism spectrum disorders. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2020;37(1):1-4.
[Crossref] [Google Scholar] [PubMed]
 Altered group and
Altered group and  Unaltered group
Unaltered group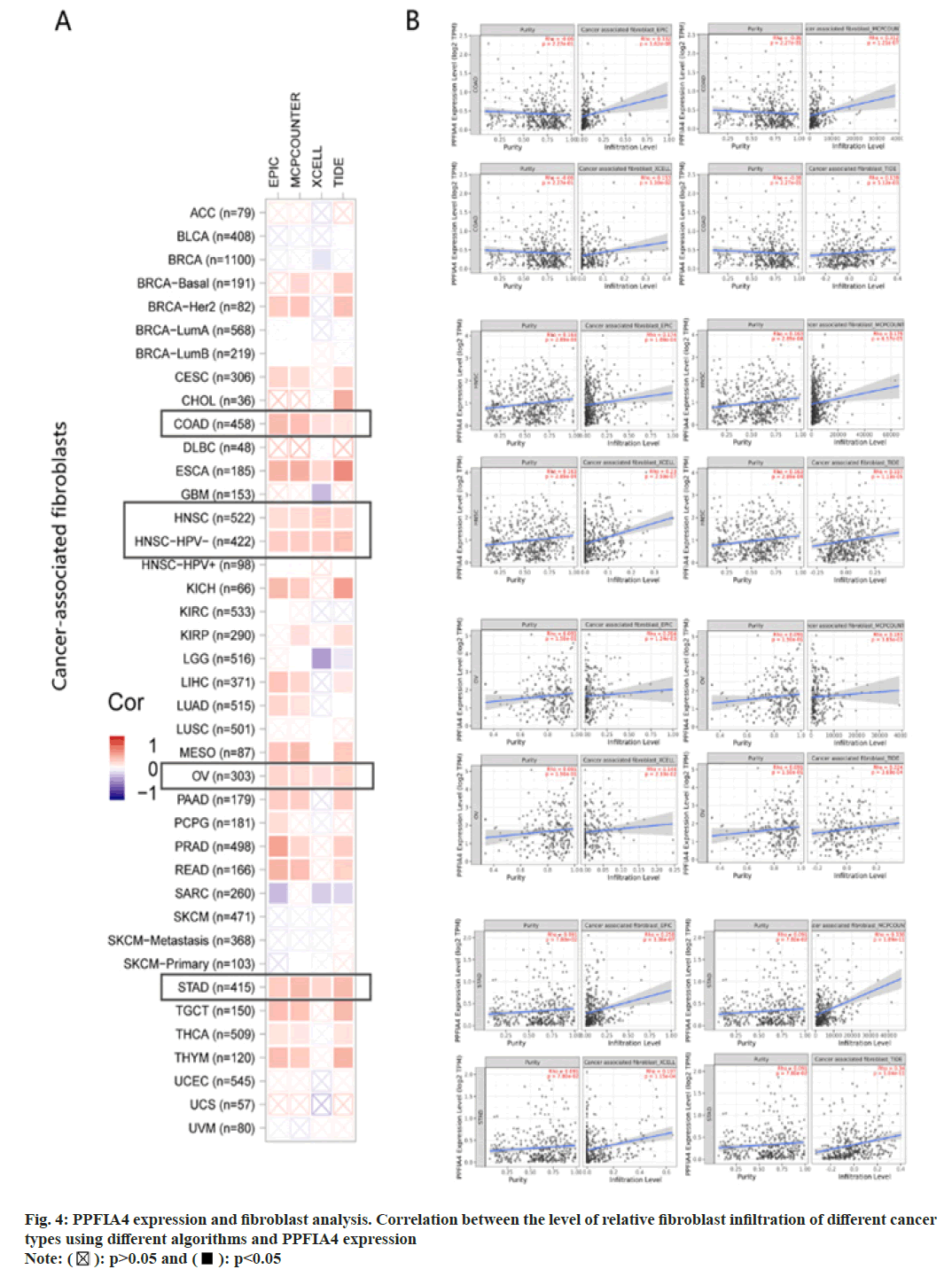
 p<0.05
p<0.05