- *Corresponding Author:
- Mulin Liu
Department of Rheumatology and Immunology, Arthritis Research Institute, the First Affiliated Hospital of Anhui Medical University, Anhui Province, Hefei 230031, China
E-mail: aydyx@163.com
| Date of Received | 10 January 2024 |
| Date of Revision | 22 July 2024 |
| Date of Accepted | 19 November 2024 |
| Indian J Pharm Sci 2024;86(6):1979-1986 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
This study aims to investigate the antimicrobial effects of eel antimicrobial peptides against Escherichia coli and Staphylococcus aureus, as well as to assess their impact on wound healing in a rat model. The antimicrobial activity was evaluated using the agar plate method, a widely accepted technique for assessing the susceptibility of bacteria to antimicrobial agents. A rat wound model was utilized to study the potential promoting effects of eel antimicrobial peptides on the wound healing process. The findings indicated that eel antimicrobial peptides not only exhibited a remarkable antimicrobial activity against the tested bacteria but also significantly accelerated the wound healing process in the rat model. The treated wounds showed faster closure, enhanced re-epithelialization, and increased collagen synthesis compared to the control group. Moreover, the eel antimicrobial peptides seemed to modulate the local immune response, promoting an environment conducive to healing without causing significant inflammation. The study also explored the possible mechanisms underlying the wound healing effects of eel antimicrobial peptides. It was hypothesized that eel antimicrobial peptides might stimulate the migration and proliferation of fibroblasts and keratinocytes, key cell types in tissue repair. Additionally, the peptides could potentially regulate the expression of growth factors and cytokines that are critical in the wound healing cascade.
Keywords
Eel antimicrobial peptides, antimicrobial activity, wound healing, Escherichia coli, Staphylococcus aureus, rat modelAm qui ommoluptae mi, eossus earcid ma nonsed molecus animus, eum
In modern medicine, the misuse of antibiotics and the increase in bacterial resistance have become global health issues. Therefore, finding new antimicrobial strategies and alternatives has become particularly important. Antimicrobial Peptides (AMPs) are a class of small molecular peptides with broad antimicrobial activity, which are widely present in the immune systems of various organisms in nature, possessing rapid and potent antimicrobial properties, and are less likely to cause bacterial resistance[1-6]. The eel, as an aquatic animal, contains a variety of bioactive substances. Recent studies have found that eel AMPs exhibit significant antimicrobial activity, capable of combating a variety of pathogenic microorganisms[3]. This study focuses on the antimicrobial effects of eel AMPs on two common pathogenic bacteria Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus). E. coli is a common intestinal bacterium, but it can cause serious infections under certain conditions, such as urinary tract infections and septicemia[5]. S. aureus, on the other hand, is a common pathogen of skin and soft tissue infections, and its drug-resistant strain Methicillin-Resistant S. aureus (MRSA) poses a huge threat to public health. Therefore, studying the antimicrobial effects of eel AMPs on these 2 bacteria has important clinical significance.
In addition to their antimicrobial effects, AMPs have also been found to have the potential to promote wound healing. Wound healing is a complex biological process involving multiple stages such as inflammatory response, cell proliferation, migration, and matrix synthesis. AMPs can reduce wound infection through their antimicrobial properties and may also directly promote the wound healing process through other mechanisms[7-9]. However, the mechanism of eel AMPs in wound healing is currently unclear and requires further research to elucidate[8].
Materials and Methods
The extraction and purification of eel AMPs:
This study initially involves the collection of samples from eels, ensuring that all samples are derived from healthy adult eels. Low-temperature freezing technology is employed for sample preservation to maintain their bioactivity. The extraction process includes tissue homogenization, multiple rounds of centrifugation, and ultrafiltration steps to remove high molecular weight impurities. Subsequently, the samples undergo purification through ion-exchange chromatography and Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC) to obtain highly pure eel AMPs. The purified AMPs are identified by mass spectrometry analysis and their amino acid sequences are determined to ensure the consistency and purity of the obtained peptide segments.
Bacterial culturing and antimicrobial activity assay:
Medium preparation: To prepare Lysogeny Broth (LB) medium, measure 100 ml of distilled water using a volumetric flask and transfer it to a 250 ml reagent bottle. Weigh 2.5 g of LB powder using an analytical balance, then add it to the water and mix thoroughly. Sterilize the mixture by autoclaving at 121° for 15 min, and set aside for use.
To prepare LB agar medium, measure 100 ml of distilled water using a volumetric flask and transfer it to a 250 ml reagent bottle. Weigh 2.5 g of LB broth powder and 1.5 g of agar powder using an analytical balance, then add both to the water and mix thoroughly. Sterilize the mixture by autoclaving at 121° for 15 min. Once the medium has cooled to approximately 40°-50°, use an electric pipette to transfer 15 ml of the medium into a disposable sterile petri dish. Take three 12 ml bacterial culture tubes and add 3 ml of LB medium to each. Inoculate each with a single colony from S. aureus (American Type Culture Collection (ATCC) code: ATCC29213) and E. coli (ATCC code: ATCC25922) solid agar plates, with one tube serving as a blank control. Place the tubes in a constant temperature shaker incubator (37°, 200 revolutions per minute (rpm)) for overnight shaking culture (15 h). The sample does not need to be sterilized and weigh a certain amount of the sample and set it aside. Dilute the bacterial solution with sterile saline to a concentration of 106 Colony Forming Unit (CFU)/ml. Add the diluted bacterial solution to the samples to achieve final concentrations of 2 mg/ml (low), 5 mg/ml (medium), and 20 mg/ml (high), with the Control group (group C) (0) receiving no sample. Place in a 37° constant temperature incubator for static culture for 48 h. After the culture is complete, make a serial 10-fold dilution of the bacterial solution with sterile Phosphate-Buffered Saline (PBS) solution, and spread 100 μl of the diluted solution evenly on the LB agar medium. Incubate at 37° for 18 h, then take photos and record the number of colonies, and further calculate the antibacterial rate of the treated groups. The colony count is the result of counting colonies at the corresponding dilution factor for each group, based on the national standard GB4789.2-2022, and colonies are counted when the number is between 30 CFU and 300 CFU. If no colonies grow at the minimum dilution factor, it proves that the sample has a significant antibacterial effect. The calculation method for bacterial solution concentration (CFU/ml) is,
Colony count×dilution factor×10 (0.1 ml spread). The antibacterial rate is calculated as,
(1-(experimental group bacterial concentration/control group bacterial concentration))×100 %.
In vitro scratch assay:
Fibroblast cells are seeded in Dulbecco's Modified Eagle Medium (DMEM), a medium containing 10 % fetal bovine serum. The cells are plated in 6-well plates and cultured in a constant temperature incubator at 37° with 5 % CO2, while ensuring the humidity is maintained at approximately 95 % to simulate the in vivo environment. Prior to seeding, the appropriate cell density and volume must be calculated based on the growth characteristics of the cells. Once the cells have formed a monolayer, the medium is replaced every 2 to 3 d to remove metabolic waste and replenish nutrients.
Before the experiment, prepare antimicrobial peptide solutions of different concentrations (0.2 %, 0.5 % and 2 %) and ensure they are mixed uniformly with the culture medium. A scratch is made on the cell monolayer using a 200 μl pipette tip, and then the plate is gently shaken to remove cellular debris and non-adherent cells from the scratch. Subsequently, the culture medium containing AMPs is added to the plate, while the group C receives an equal volume of medium without the peptides. At 0 h, 12 h, 24 h, and 72 h post-scratch, observe the cell morphology and scratch healing using an inverted microscope, and capture images of the scratched area with a time-stamped camera. These images will be used for subsequent analysis of the healing effect. To calculate the wound healing rate, measure the length and width of the wound at different time points using image analysis software to determine the wound area. The healing rate (%) is then calculated using the formula,
Healing rate (%)=[(1-(wound Area at time T/original wound area))×100 %].
Establishment and treatment of a rat wound model:
Select 32 healthy Sprague Dawley (SD) rats, aged 6-8 w and weighing between 180-220 g, as experimental animals (production certificate: SCXK (Jing) 2019- 0010). The animals are housed in a barrier system laboratory with controlled conditions (air cleanliness ≤10,000 grade, air exchange rate 10-20 times per hour, temperature 20°-26°, daily temperature variation ≤3°, relative humidity 40 %-70 %), and provided with a 12 h light and 12 h dark cycle. All animals are adaptively fed for 1 w with ample rodent chow and water. Establish a full thickness skin wound model. Under sterile conditions, a standardized incision (approximately 2 cm) is made on the back of the rat to create a wound of a certain area. The purified eel AMPs are applied topically to the wound surface at concentrations of 0.2 % (Low dose group (group L)), 0.5 % (Medium dose group (group M)), and 2 % (group H) in physiological saline solution, while the group C is treated with physiological saline. The day the model is established is designated as day 0, and treatment according to the experimental design begins for each group. The medication is applied to cover the wound, and then the wound is sealed with 3M™ Tegader™-film transparent dressing. Photographs are taken to calculate the healing rate and wound area on 0, 3, 6, 9, 12, and 14 d. All animals are raised in a standardized environment, and the wound healing process is regularly monitored.
Data collection and statistical analysis methods:
The collected data includes macroscopic observations of wound healing, microscopic structural changes in wound tissue, and bacterial growth curves. Quantitative analysis of wound healing is performed using image analysis software. All experiments are independently repeated at least three times to ensure the reliability and reproducibility of the data. Data analysis is conducted using statistical software, including t-tests and Analysis of Variance (ANOVA), to assess whether the differences between the experimental and control groups are statistically significant. A p-value <0.05 is considered to indicate a significant difference.
Results and Discussion
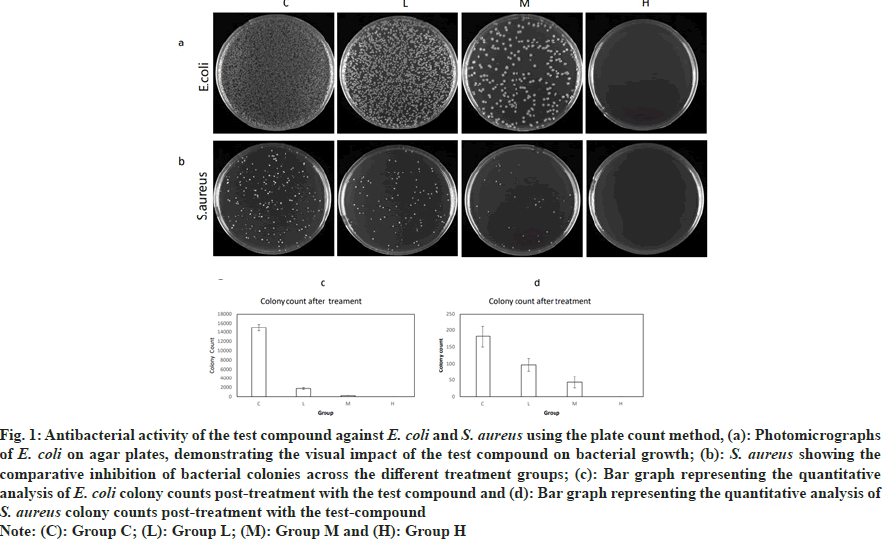
The purity of the AMPs was determined as 98.7 % and the sequence was identified as RWKIFKKIEKVGQNVRDYIKFLR. In control group the colony count of E. coli was 15 048 and the colony count of S. aureus was 182. In low dose group the colony count of E. coli was 1753 with a bacterial inhibition rate of 88.35 %; the colony count of S. aureus was 97, with a bacterial inhibition rate of 46.70 %. In group M the colony count of E. coli was 227, with a bacterial inhibition rate of 98.49 %; the colony count of S. aureus was 43, with a bacterial inhibition rate of 76.37 %. In group H the colony count of E. coli was 0, with a bacterial inhibition rate of 99.99 %; the colony count of S. aureus was 0, with a bacterial inhibition rate of 99.45 %. The experimental results indicate that as the concentration of the antimicrobial peptide increases, the antibacterial effect on E. coli and S. aureus significantly strengthens. The bacterial inhibition rate of the group H is much higher than that of the low and group M, demonstrating that the antimicrobial peptide has a concentration-dependent antibacterial effect (fig. 1).
Fig. 1: Antibacterial activity of the test compound against E. coli and S. aureus using the plate count method, (a): Photomicrographs
of E. coli on agar plates, demonstrating the visual impact of the test compound on bacterial growth; (b): S. aureus showing the
comparative inhibition of bacterial colonies across the different treatment groups; (c): Bar graph representing the quantitative
analysis of E. coli colony counts post-treatment with the test compound and (d): Bar graph representing the quantitative analysis of
S. aureus colony counts post-treatment with the test-compound
Note: (C): Group C; (L): Group L; (M): Group M and (H): Group H
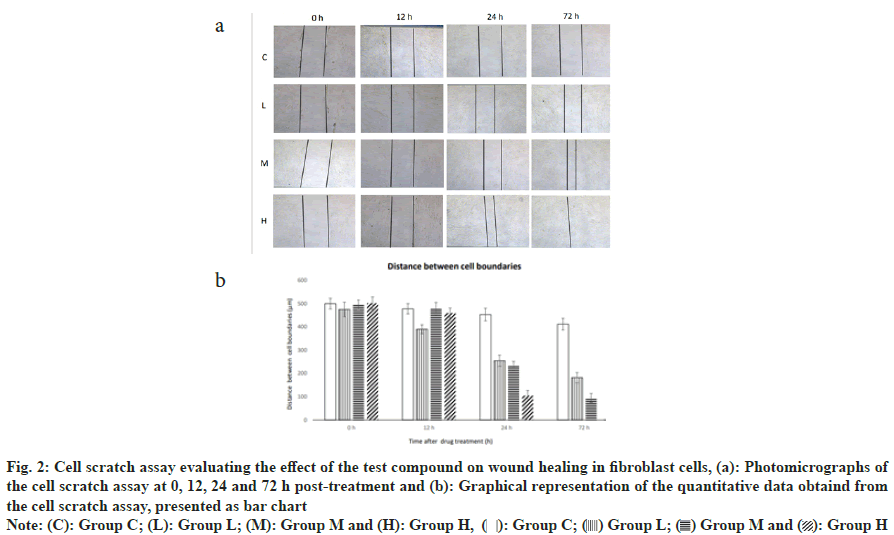
At the start of the experiment, the wound width was consistent across all groups, measuring approximately 500 μm. After 12 h, in the group C, cells migrated to the edge of the wound, reducing the width to 478 μm. In the low dose antimicrobial peptide group, cell migration was slightly faster, reducing the wound width to 390 μm. The medium dose antimicrobial peptide group showed no significant change, with the wound width remaining at 480 μm, while the high dose antimicrobial peptide group had a reduction to 460 μm. At 24 h, the wound width in the group C, was 453 μm, while in the low dose antimicrobial peptide group it was reduced to 255 μm, in the medium dose antimicrobial peptide group it was reduced to 235 μm, and in the high dose antimicrobial peptide group it was reduced to 105 μm. At 72 h, the wound width in the group C was 412 μm, while in the low dose antimicrobial peptide group it was reduced to 182 μm, in the medium dose antimicrobial peptide group it was reduced to 95 μm, and in the high dose antimicrobial peptide group, the cells exhibited the best quality of healing, with the wound nearly fully healed. The antimicrobial peptide demonstrated an effect in promoting the migration of fibroblasts and wound healing at low, medium, and high doses. With the increase in the dose of the antimicrobial peptide, both the speed of cell migration and the quality of healing were improved. The group H nearly completed the wound healing within 72 h, showing a significant effect in promoting wound healing. The medium and high dose groups had better healing quality than the group C at 72 h, indicating that the antimicrobial peptide may have a positive effect on the reconstruction of the extracellular matrix and tissue repair (fig. 2).
Fig. 2: Cell scratch assay evaluating the effect of the test compound on wound healing in fibroblast cells, (a): Photomicrographs of
the cell scratch assay at 0, 12, 24 and 72 h post-treatment and (b): Graphical representation of the quantitative data obtaind from
the cell scratch assay, presented as bar chart
Note: (C): Group C; (L): Group L; (M): Group M and (H): 
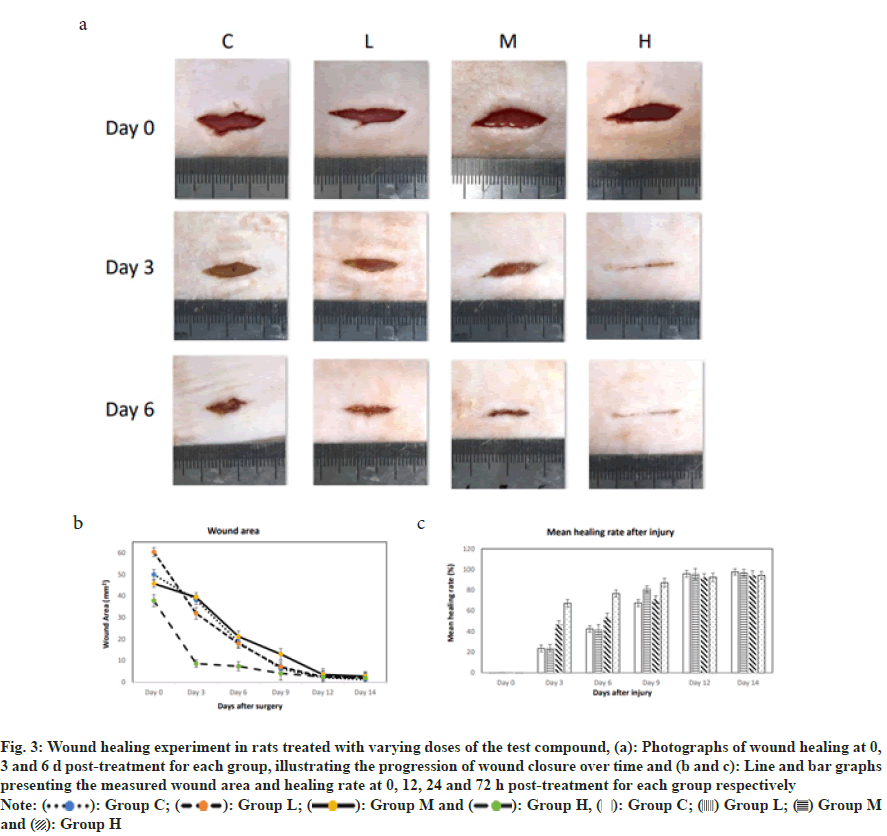
The wound area in all groups significantly decreased over time, indicating the wound healing process. At all-time points, the rate of wound healing in the blank group C was lower than that of the other three groups, suggesting that the test substance promotes wound healing.
The rate of wound healing increased with the dose of the test substance. Specifically, in the group L, the wound area decreased from 60.3 mm2 on 0th d to 2 mm2 on 14th d; in the group M, the wound area decreased from 45.7 mm2 on 0th d to 2.7 mm2 on 14th d; and in the group H, the wound area decreased from 37.8 mm2 on 0th d to 1.8 mm2 on 14th d. The wound healing rate in all groups significantly increased over time, indicating the wound healing process. At all-time points, the healing rate of the blank group C was lower than that of the other three groups, suggesting that the test substance promotes wound healing. The wound healing rate correspondingly increased with the increase in the dose of the test substance. Specifically, in the group L, the wound healing rate increased from 0.3 % on 0th d to 96.7 % on 14th d; in the group M, the wound healing rate increased from 0.3 % on 0th d to 94.1 % on 14th d; and in the group H, the wound healing rate increased from 0.2 % on 0th d to 94.3 % on 14th d.
The wound healing rate in all groups showed a trend of accelerating over time. Particularly, the most significant increase in healing rate occurred between 3rd d and 6th d post-treatment, which may be the critical period for the test substance's effect on wound healing. In conclusion, the test substance significantly promotes the wound healing process in animals, and its effect becomes more pronounced with increasing doses. The fastest rate of wound healing occurred between 3rd d and 6th d post-treatment, indicating this period may be the key window for the drug's action (fig. 3).
Fig. 3: Wound healing experiment in rats treated with varying doses of the test compound, (a): Photographs of wound healing at 0,
3 and 6 d post-treatment for each group, illustrating the progression of wound closure over time and (b and c): Line and bar graphs
presenting the measured wound area and healing rate at 0, 12, 24 and 72 h post-treatment for each group respectively
 and
and 
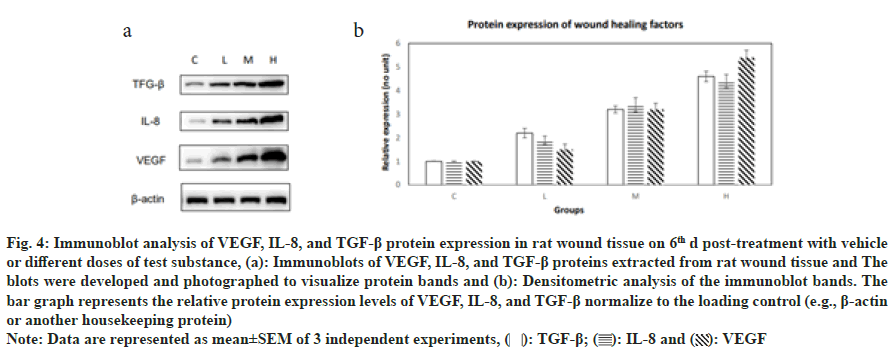
In this study, we evaluated the relative expression levels of proteins known to promote wound healing across different treatment groups using immunoblotting. The expression of Transforming Growth Factor-Beta (TGF-β) showed a dose-dependent increase, with the group C exhibiting the lowest level 1 and the group H showing the highest level (4.6). This suggests that the presence of the test substance significantly upregulates TGF-β expression, which is instrumental in tissue repair and wound healing processes. Interleukin-8 (IL- 8) expression levels also increased with the dose of the test substance. The group C had the lowest expression 1, and the group H had the highest expression (4.4). IL-8 is a potent chemoattractant for neutrophils and plays a critical role in the inflammatory phase of wound healing. Vascular Endothelial Growth Factor (VEGF) expression levels demonstrated a similar trend, with the lowest level in the group C and the highest level in the group H (5.4). VEGF is a key regulator of angiogenesis and is essential for the proliferative phase of wound healing (fig. 4). The results indicate a clear dose-dependent response in the expression of all three proteins, with higher doses of the test substance leading to increased expression levels. This suggests that the test substance may enhance wound healing by modulating the expression of these critical proteins involved in the wound healing cascade.
Fig. 4: Immunoblot analysis of VEGF, IL-8, and TGF-β protein expression in rat wound tissue on 6th d post-treatment with vehicle
or different doses of test substance, (a): Immunoblots of VEGF, IL-8, and TGF-β proteins extracted from rat wound tissue and The
blots were developed and photographed to visualize protein bands and (b): Densitometric analysis of the immunoblot bands. The
bar graph represents the relative protein expression levels of VEGF, IL-8, and TGF-β normalize to the loading control (e.g., β-actin
or another housekeeping protein)
Note: Data are represented as mean±SEM of 3 independent experiments, 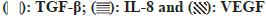
In this study, we systematically investigated the antimicrobial effects of eel AMPs (EAMPs) against E. coli and S. aureus, as well as their influence on the wound healing process in a rat model. The findings from the antibacterial assays and cell scratch assays provide compelling evidence that EAMPs possess potent antimicrobial activity and can significantly accelerate wound closure[6]. The results from the antimicrobial assays clearly demonstrate a concentration-dependent bactericidal effect of EAMPs on both E. coli and S. aureus. The group H exhibited nearly complete inhibition of bacterial growth, underscoring the peptides robust antimicrobial potential[2]. This observation is consistent with the premise that AMPs can disrupt bacterial cell membranes and inhibit essential cellular processes, leading to cell death. The differential sensitivity of the two bacterial species to EAMPs may be attributed to variations in cell wall composition and structure, which is a common theme in antimicrobial research[7]. The cell scratch assay results corroborate the in vitro antimicrobial activity with an in vivo wound healing application. The increased rate of wound closure in response to EAMPs treatment suggests that these peptides may not only prevent infection but also actively promote tissue repair. The acceleration in wound healing, particularly in the group H, indicates that EAMPs could be harnessing the body's innate regenerative mechanisms, potentially by modulating cytokine expression and enhancing cell migration and proliferation. The mechanistic study provides intriguing insights into how EAMPs might be influencing the wound healing process. The upregulation of TGF-β, IL-8, and VEGF in a dose-dependent manner indicates that EAMPs could be mediating their effects through known pathways involved in tissue repair and angiogenesis. TGF-β is a multifunctional cytokine that plays a central role in wound healing by regulating cell proliferation, differentiation, and extracellular matrix production. Similarly, IL-8 is a key chemokine involved in neutrophil chemotaxis, which is essential for the initial inflammatory response to injury. VEGF's role in promoting angiogenesis is well-established, and its upregulation in response to EAMPs suggests an enhancement of the wound's vascularization, a critical component of the healing process[4].
The findings from this study have significant clinical implications, particularly in the context of chronic wounds and antibiotic-resistant infections. EAMPs could offer a novel therapeutic strategy that combines antimicrobial action with wound healing promotion. Future research should focus on translating these preclinical results into clinical applications, including the development of EAMPs-based wound care products and the assessment of their safety and efficacy in human subjects. Moreover, understanding the precise molecular mechanisms by which EAMPs enhance wound healing could lead to the discovery of new targets for therapeutic intervention. The potential of EAMPs to modulate the immune response and promote tissue regeneration warrants further investigation, particularly in the context of their impact on various stages of the wound healing process[9]. In conclusion, this study provides robust evidence that EAMPs exhibit significant antimicrobial activity against both E. coli and S. aureus in a dose-dependent manner. Furthermore, EAMPs demonstrate the potential to accelerate wound healing in a rat model by promoting cell migration and modulating the expression of key proteins involved in tissue repair. These findings suggest that EAMPs could represent a novel therapeutic strategy for treating infections and enhancing wound recovery, warranting further investigation for clinical applications[10].
Authors’ contributions:
Haigang Cui served as the corresponding author for this study, overseeing the project's direction, providing critical feedback on the research design, and finalizing the manuscript for submission. His expertise and guidance were instrumental in ensuring the accuracy and integrity of the research findings.
Mulin Liu, as the first author, took the lead in conducting the experiments, analyzing the data, and drafting the initial manuscript. Her dedication to the research process and meticulous attention to detail contributed significantly to the quality of the work presented. Each author has contributed to the work in a meaningful way, and all have approved the final version of the manuscript.
Acknowledgements:
The authors would like to express their sincere gratitude to all individuals who contributed to the success of this research project. We extend our special thanks to Haigang Cui, the corresponding author, for his invaluable guidance and mentorship throughout the project. His expertise and insights have been pivotal in shaping the research direction and ensuring the quality of our work. Our appreciation goes to Mulin Liu, the first author, for her tireless efforts in conducting the experiments, analyzing the data, and drafting the manuscript. Her commitment and attention to detail have been fundamental to the advancement of our research. We are also grateful to our colleagues and collaborators for their support and constructive feedback during the various stages of the project. Their contributions have significantly enhanced the depth and breadth of our study. Lastly, we thank our families for their understanding and encouragement, providing us with the emotional support necessary to complete this demanding project.
Data availability statement:
The data that support the findings of this study are available upon request. The corresponding author, Haigang Cui, can be contacted for all data-related inquiries. The raw data and processed data sets generated during the current study are not publicly available due to proprietary restrictions. However, de-identified data can be made available for researchers who meet the criteria for access to confidential data.
Ethical statement:
The study was conducted in accordance with the Chinese Principles of the Laboratory Animal Guideline for Ethical Review of Animal Welfare No. GB/T 35892- 2018.
Funding:
This study was funded by Ganglu Biotech grant No. GL20240025.
Conflict of interest:
The authors declared no conflict of interests.
References
- Boman HG. Peptide antibiotics and their role in innate immunity. Ann Rev Immunol 1995;13(1):61-92.
[Crossref] [Google Scholar] [PubMed]
- Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 2005;3(3):238-50.
- Chan, BY, Fjell CD. Design principles for antimicrobial peptides: A case for optimization. Biochem J 2017;474(2):219-34.
- Dathe M, Wieprecht T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim Biophys Acta 1999;1462(1-2):71-87.
[Crossref] [Google Scholar] [PubMed]
- Gough M, Davidson DJ. Antimicrobial activity of modified host defense peptides against clinical isolates of Staphylococcus aureus andPseudomonas aeruginosa. J Antimicrob Chemother 2008;61(5):1168-75.
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 2006;24(12):1551-7.
- Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev 2006;19(3):491-511.
- Massin M, Von Bernuth G. Clinical and haemodynamic correlates of heart rate variability in children with congenital heart disease. Eur J Pediatr 1998;157:967-71.
[Crossref] [Google Scholar] [PubMed]
- Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol 2011;29(9):464-72.
- Zasloff M. Antimicrobial peptides in innate immunity: From mollusks to mammals. Nat Rev Microbiol 2002;2(4):529-38.