- *Corresponding Author:
- Yan Jin
Department of Clinical Oncology, The Affiliated Huaian No.1 People’s Hospital, Nanjing Medical University, Huaian, Jiangsu 223300, China
E-mail: jinyan_05170422@126.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “217-225” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Emerging evidence has revealed that promyelocytic leukemia zinc finger may contribute in several ways to the development and progression of cancer, but no pan-cancer analysis of its roles have been undertaken to date. Here, we set out to use bioinformatics techniques to comprehensively investigate the expression profiles and prognostic significance of promyelocytic leukemia zinc finger, as well as its relationship to clinic pathological parameters and immune cell infiltration. Promyelocytic leukemia zinc finger is significantly down regulated in various cancers, which has been found to be associated with patient prognosis. We also found that the under expression of promyelocytic leukemia zinc finger is often associated with poor overall survival and disease-specific survival in chromophobe renal cell carcinoma and renal clear cell carcinoma. Remarkably, promyelocytic leukemia zinc finger expression has been found to correlate with levels of infiltrating cells, the tumor mutational burden and microsatellite instability. Our pan-cancer analysis thus provided a deeper understanding of the functions of promyelocytic leukemia zinc finger in oncogenes and the metastasis of different cancers.
Keywords
Pan-cancer analysis, promyelocytic leukemia zinc finger, oncogenes, tumor, immunotherapy
Cancer is one of the major causes of death and the biggest barrier to the extension of life expectancy worldwide. In 2020, approximately 19.3 million new cancer cases and 10 million cancer-related deaths were recorded globally[1]. An increasing body of evidence demonstrates that the occurrence and development of cancer is closely related to abnormalities in genes. Thus, further analysis of alterations in oncogenes and tumor suppressor genes may help to identify cancer and cancer-related mechanisms by uncovering prognostic relationships which can be used to inform clinical practice[2,3]. Though recent advancements in targeted and immune-modulating therapies have improved the odds of survival for many cancer patients[4], treatment of cancer is still unsatisfactory and the pathogenesis of cancer to identify potential therapeutic targets urgently requires further exploration.
The zinc finger protein family is considered one of the most abundant families of regulatory proteins in eukaryotic cells, with more than 200 known members. Zinc Finger and BTB Domain Containing 16 (ZBTB16), also known as Promyelocytic Leukemia Zinc Finger (PLZF), was first discovered in acute promyelocytic leukemia in the form of a fusion protein with the Retinoic Acid Receptor Alpha (RARA)[5,6]. PLZF-RAR fusion protein inhibits the normal function of PLZF and RARA, which has been shown to contribute to the occurrence and development of acute promyelocytic leukemia. Recent studies have demonstrated that PLZF is involved in the occurrence, development and prognosis of solid cancers and overexpression of PLZF messenger Ribonucleic Acid (mRNA) has been shown to suppress gall bladder cancer proliferation in vitro and in vivo[7]. On the other hand, decreases in PLZF expression have been shown to promote the development of an enzalutamide-resistant phenotype in prostate cancer cells[8]. PLZF is down regulated in the majority of gastric cancer cell lines and tumor tissues. The down regulation of PLZF has been closely associated with high-risk clinical stages and metastasis in gastric cancer, which is indication of poor prognosis[9]. The biological functions of PLZF have been extensively examined. However, few studies have comprehensively explored the specific roles of PLZF across various cancer types.
In this study, we set out to conduct a comprehensive analysis of this topic based on data from several databases. We visualized the expression and prognostic value of PLZF across different cancer types, investigated its relationship to immunotherapy response and then considered possible molecular biological functions. We found that PLZF was under expressed in multiple cancers, which was associated with patient prognosis. Overall, our research demonstrated the important role of PLZF in anti-tumor immunity and immunotherapy response, potentially providing new targets for cancer treatment.
Materials and Methods
Gene expression analysis:
Using Tumor Immune Estimation Resource (TIMER), version 2 (http://timer.comp-genomics.org/), we analyzed the expression of PLZF, mRNA in various cancers. Differential expression of PLZF between normal and cancerous tissue was observed for various types cancers, and the range was set as follows: log2 fold change (log2 FC) ≥1 or ≤-1 and p≤0.05. In addition, Gene Expression Profiling Interactive Analysis (GEPIA), version 2, was employed to analyze PLZF, mRNA levels in certain tumors where there was no corresponding normal tissue[10]. The University of Alabama at Birmingham Cancer Data Analysis Portal (UALCAN), an interactive and synthetic online platform made to aid in the analysis of open-source. The Cancer Genome Atlas (TCGA) data, was used to analyze expression levels of proteins with the confirmatory/ discovery tool of the Clinical Proteomic Tumor Analysis Consortium (CPTAC). Additionally, violin plots were constructed to visualize the relationship between PLZF and cancer pathological stages by using the “stage plot” module of GEPIA2.
Prognosis and survival analysis:
A “survival map” was developed to detect the Overall Survival (OS) and the Disease-Free Survival (DFS) of PLZF in all the tumors in TCGA data. The values used to divide groups with high and low expressions were defined as the cutoff-high (50 %) and cutoff-low (50 %) values, respectively. Survival plots were obtained using the “survival analysis” module of GEPIA2, while survival rates between different groups were compared using log-rank tests.
Analysis of genetic alterations:
Data about PLZF genetic mutation type, alteration frequency, mutated sites and copy number were retrieved through cBioPortal. Information on difference between cases of OS, DFS and Progression-Free Survival (PFS) that occurred within the same cancer type with and without PLZF alteration was obtained using the “comparison” module. Kaplan–Meier plots with associated log-rank p values were also generated.
Gene Set Enrichment Analysis (GSEA):
To study the potential roles of PLZF in cancer, we divided samples into high and low expression groups based on the PLZF expression of each cancer type. High and low expression groups consisted of the top and bottom 30 % of samples displaying PLZF expression, respectively. GSEA (http://www.gseamsigdb.org/gsea/index.jsp) was then performed using the “clusterProfiler” R package (v4.2.0). PLZF is a key regulator of the innate T cell lineage. To analyze this, GSEA gene sets were downloaded from the molecular signatures database (v7.5.1). A False Discovery Rate (FDR) adjusted, p<0.05 threshold was used to define statistical significance.
Analysis of the correlation between PLZF expression and tumor cell infiltration:
Immune checkpoint inhibitor is a promising therapy for tumor immunotherapy[11]. To analyze the immunological roles of PLZF, including its correlation with diverse immune cells and immune regulation, we employed the TIMER tool. A Spearman correlation analysis heat map of the immune score or immune checkpoint-related genes and PLZF gene expression in multiple cancers was also generated. Various types of cancer were represented on the horizontal axis of the heat maps, the vertical axis represented the immune scores and different colors represented correlation coefficients. R software v4.2.0 was used for statistical analysis (*p<0:05, **p<0:01 and ***p<0:001).
Analysis of the relationship between PLZF gene expression and Tumor Mutational Burden (TMB) or Microsatellite Instability (MSI):
TMB and MSI scores were isolated from the TCGA data. Correlation analysis between PLZF expression and TMB or MSI was then performed using Spearman’s method. The horizontal axis in the figure represents the correlation coefficient between PLZF and TMB or MSI. Here, the ordinate indicates different types of cancer, while the size of the dots represents the size of the correlation coefficient. Colors represent significance of p values.
PLZF-binding proteins enrichment analysis:
PLZF-binding proteins were located using the STRING website with the query “PLZF”. The Pearson correlation coefficient was calculated to analyze the first 100 PLZFinteracting genes based on differential expression levels in TCGA tumors and normal tissues using GEPIA2. A heat map consisting of the top 5 genes found through this process and containing the partial correlation index was constructed, while p values were obtained using the “Genecorr” component of TIMER2 using the purity-adjusted Spearman’s rank correlation test. A Venn diagram viewer allowed for the visualization of PLZF-binding and interacting protein genes. Two sets of data were combined to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Results and Discussion
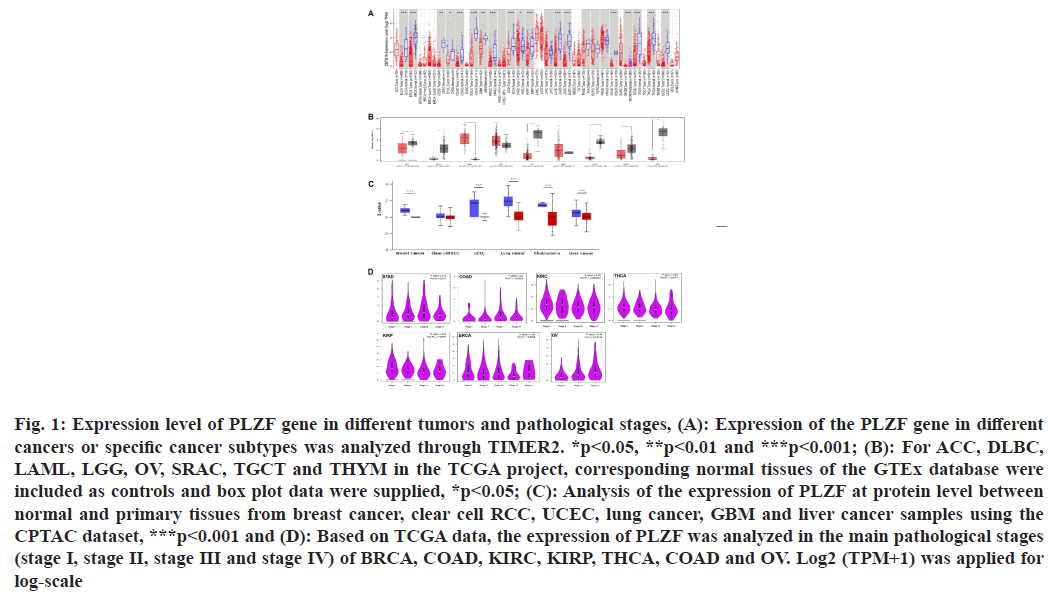
We first assessed PLZF expression levels in different cancers. PLZF expression was lower in Bladder Urothelial Carcinoma (BLCA) (p<0.001), Breast Invasive Carcinoma (BRCA) (p<0.001), Cervical Squamous Cell Carcinoma (CESC) (p<0.01), Cholangiocarcinoma (CHOL) (p<0.05), Colon Adenocarcinoma (COAD) (p<0.001), Esophageal Carcinoma (ESCA) (p<0.001), Glioblastoma Multiforme (GBM) (p<0.01), Head and Neck Squamous Cell Carcinoma (HNSC) (p<0.001), Kidney Chromophobe (KICH) (p<0.001), Kidney Renal Clear Cell Carcinoma (KIRC) (p<0.05), Kidney Renal Papillary Cell Carcinoma (KIRP) (p<0.001), Lung Adenocarcinoma (LUAD) (p<0.001), Lung Squamous Cell Carcinoma (LUSC) (p<0.001), Rectum Adenocarcinoma (READ) (p<0.001), Stomach Adenocarcinoma (STAD) (p<0.001) and Uterine Corpus Endometrial Carcinoma (UCEC) (p<0.01) than in tumor-adjacent tissues. However, no differential expression of PLZF was observed between Liver Hepatocellular Carcinoma (LIHC), Pancreatic Adenocarcinoma (PAAD), Pheochromocytoma and Paraganglioma (PCPG) and Prostate Adenocarcinoma (PRAD) tumors and adjacent tissues (fig. 1A). We further explored whether PLZF was differentially expressed between tumor and normal tissue for cancer types where data on corresponding normal tissue was not available on TCGA. From this, we found that Adrenocortical Carcinomas (ACCs) (p<0.05), Diffuse Large B-cell Lymphomas (DLBC) (p<0.05), Acute Myeloid Leukemias (AML) (p<0.05), Ovarian Serous Cystadenocarcinomas (OV), Testicular Germ Cell Tumors (TGCT), Thymomas (THYM) and Uterine Carcinosarcomas (UCS) exhibited lower expressions of PLZF. No significant differences in PLZF expression were detected between Lower Grade Glioma (LGG) and Sarcoma (SARC) samples relative to corresponding non-cancerous tissues (fig. 1B).
Fig. 1: Expression level of PLZF gene in different tumors and pathological stages, (A): Expression of the PLZF gene in different cancers or specific cancer subtypes was analyzed through TIMER2. *p<0.05, **p<0.01 and ***p<0.001; (B): For ACC, DLBC, LAML, LGG, OV, SRAC, TGCT and THYM in the TCGA project, corresponding normal tissues of the GTEx database were included as controls and box plot data were supplied, *p<0.05; (C): Analysis of the expression of PLZF at protein level between normal and primary tissues from breast cancer, clear cell RCC, UCEC, lung cancer, GBM and liver cancer samples using the CPTAC dataset, ***p<0.001 and (D): Based on TCGA data, the expression of PLZF was analyzed in the main pathological stages (stage I, stage II, stage III and stage IV) of BRCA, COAD, KIRC, KIRP, THCA, COAD and OV. Log2 (TPM+1) was applied for log-scale
In addition to the level of transcription, we also obtained levels of PLZF expression at the protein level from the CPTAC dataset, where we found that PLZF protein levels were significantly lower in BRCA (p<0.001), UCEC (p<0.001), LUAD (p<0.001), GBM (p<0.001) and LIHC (p<0.001) than those in normal tissues (fig. 1C).
We assessed PLZF expression in patients with stage I, II, III and IV cancers to investigate the relationship between PLZF expression and clinic pathological features in various cancers. Analysis of data from the TCGA database found that the expression of PLZF was significantly decreased in BRCA (p=0.0208), COAD (p=0.0403), KIRC (p<0.001), KIRP (p=0.00779), STAD (p=0.011) and OV (p=0.0134) (fig. 1D).
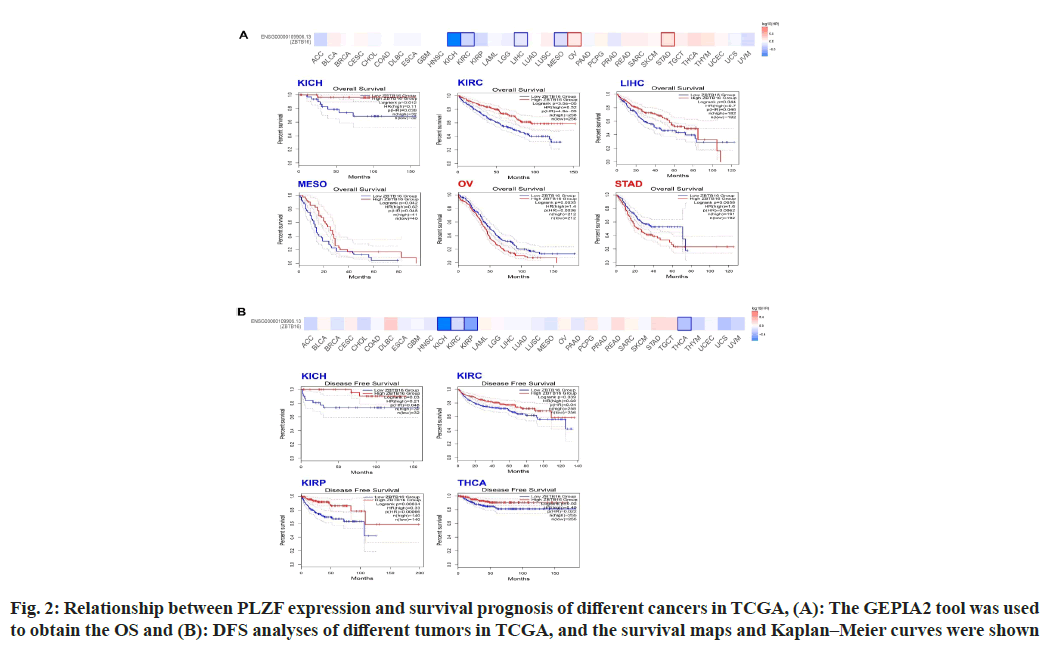
To estimate the relationship between PLZF expression and prognosis, we separated cases from the TCGA database into two groups based on PLZF expression and compared cancer outcomes across the pan-cancer dataset. Fig. 2 shows the OS and DFS for different kinds of tumors, while the survival map shows the relationship between PLZF expression and OS. We found that among cases of KICH (p=0.0012), KIRC (<0.001) and LIHC (p=0.044), higher levels of PLZF expression were associated with longer survival periods (fig. 2A). On the other hand, lower expression of PLZF in cases of OV (p=0.035) and STAD (p=0.0056) was associated with shorter survival periods (fig. 2A). Analysis of DFS data revealed that low expression of PLZF is linked to poor prognosis for KICH (p=0.03), KIRC (p=0.039), KIRP (p<0.001) and Tetrahydrocannabinolic Acid (THCA) (p=0.02) (fig. 2B).
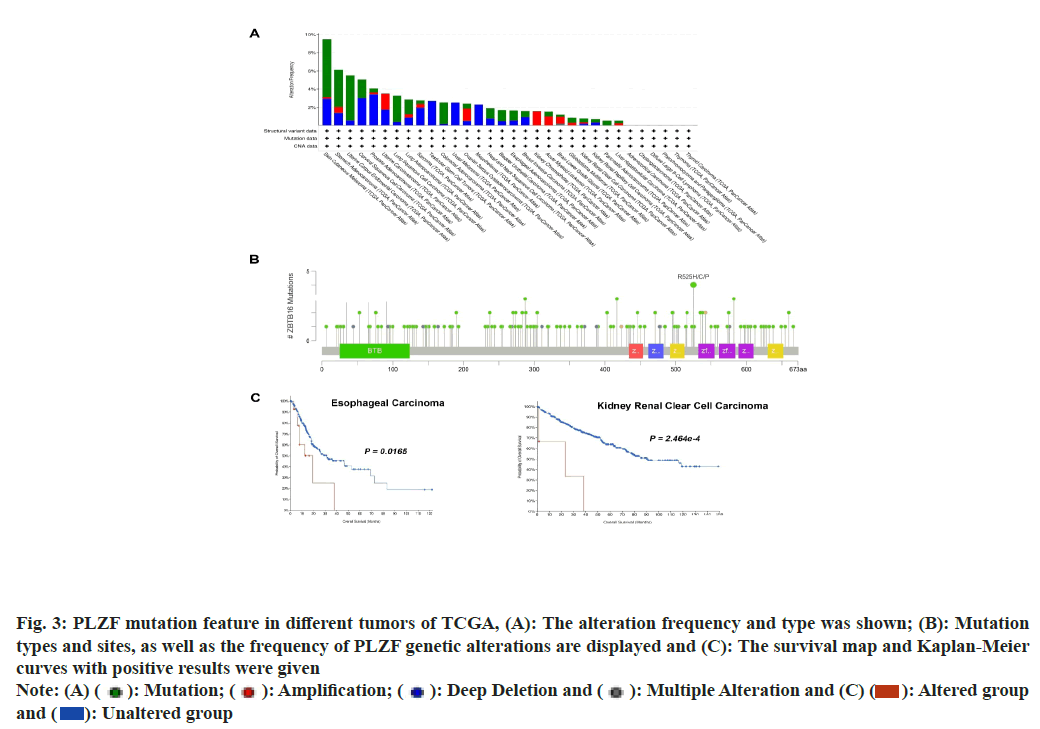
Mutations of PLZF in cancer were studied using data acquired from the cBioPortal database, which allowed the analysis of types and sites of genetic mutation of PLZF in different cancers. The frequency of PLZF mutation (>5 %) was highest in patients with Skin Cutaneous Melanoma (SKCM), STAD, UCEC and CESC (fig. 3A). In addition, the types, sites and numbers of the PLZF genetic mutations were also examined. A total of 166 PLZF mutations were identified, consisting of 141 missense, 19 truncating, 4 splice and 2 synaptic vesicle/fusion mutations. An R525H/C/P mutation was found in 2 cases of STAD, 1 case of HNSC and 1 case of LUAD (fig. 3B). We also studied the relationship between the genetic mutation of PLZF and survival prognosis relative to different types of tumors. Notably, cases of ESCA and KIRC with PLZF mutation were found to be associated with the poorest OS (fig. 3C).
Fig. 3: PLZF mutation feature in different tumors of TCGA, (A): The alteration frequency and type was shown; (B): Mutation
types and sites, as well as the frequency of PLZF genetic alterations are displayed and (C): The survival map and Kaplan-Meier
curves with positive results were given
Note: (A) ( ): Mutation; (
): Mutation; (  ): Amplification; (
): Amplification; ( ): Deep Deletion and (
): Deep Deletion and ( ): Multiple Alteration and (C) (
): Multiple Alteration and (C) ( ): Altered group
and (
): Altered group
and ( ): Unaltered group
): Unaltered group
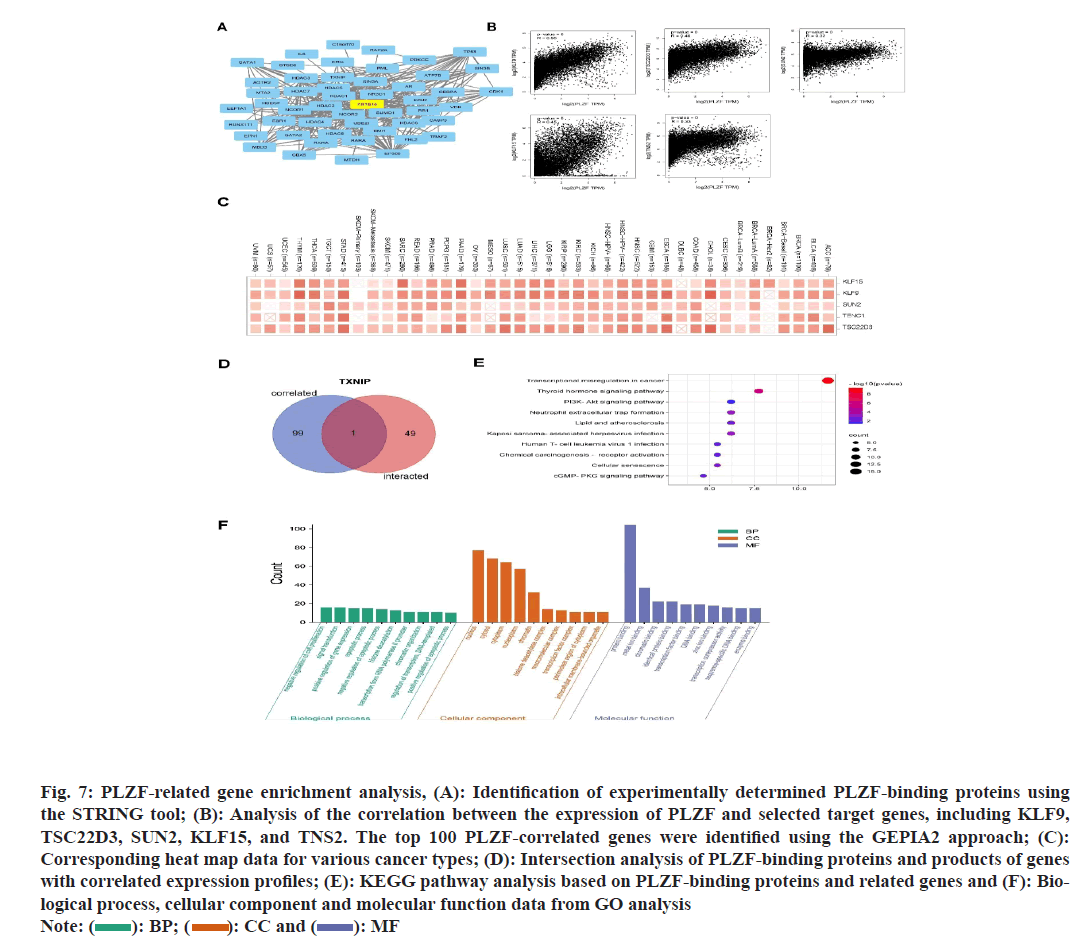
PLZF-binding proteins and PLZF-related genes were examined to better understand possible mechanisms underlying the link between PLZF function and tumor oncogenesis. Protein-Protein Interaction (PPI) networks were constructed based on 50 PLZF-binding proteins using the STRING tool (fig. 4A), after which the GEPIA2 tool was used to explore the top 100 PLZF-associated genes. As illustrated in fig. 4B, expression of PLZF was positively correlated with that of KLF9, TSC22D3, SUN2, KLF15 and TNS2 across a range of various cancers (fig. 4C). Among these, Thioredoxin Interacting Protein (TXNIP) was identified as the only protein with both PLZF-binding properties and correlated expression (fig. 4D), which promoted glucose uptake and glycolysis, induced hyper glycolytic T helper-17 (Th17) like regulatory T (Treg), facilitated Th17 inflammation, promoted interleukin 17A induced of Cluster of Differentiation 8 (CD8+) T-cell exhaustion and drove carcinogenesis[12]. KEGG and GO enrichment analyses were then performed. KEGG analysis suggested that the term “transcriptional misregulation in cancer” was associated with the role of PLZF in tumor oncogenesis (fig. 4E). By analyzing the GO enrichment data, we also found that these genes were correlated with certain processes including protein binding, metal ion binding, chromatin binding and identical protein binding and transcription factor binding (fig. 4F).
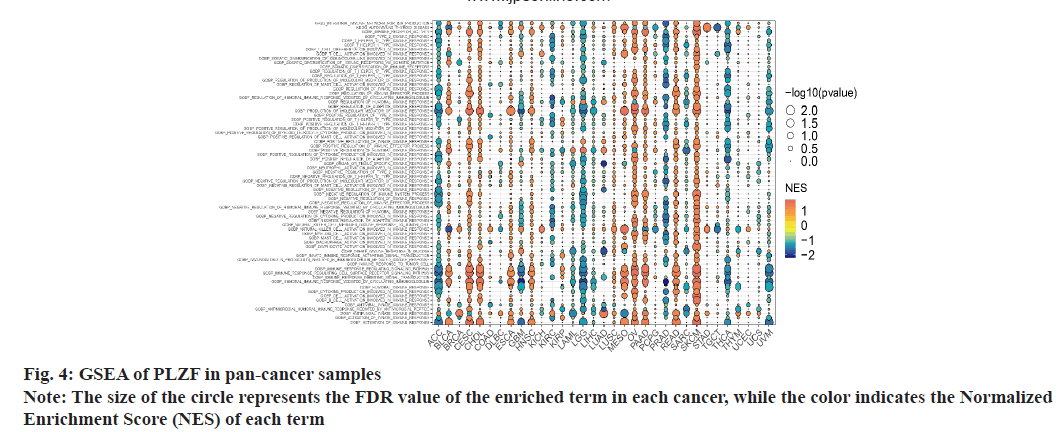
As PLZF is a key regulator of the innate T cell lineage, GSEA was performed based on findings from GO and KEGG analysis (fig. 5). In ACC, CESC, LGG, Malignant Mesothelioma (MESO), OV, PAAD, PRAD, READ, SKCM, THCA and Uveal Melanoma (UVM), we found that PLZF expression was closely related to immune-related pathways, including innate immune response, adaptive immune response, humoral immune response and inflammatory cells such as neutrophils, lymphocytes and natural killer cells. These results illustrate the important role of PLZF in immune regulation.
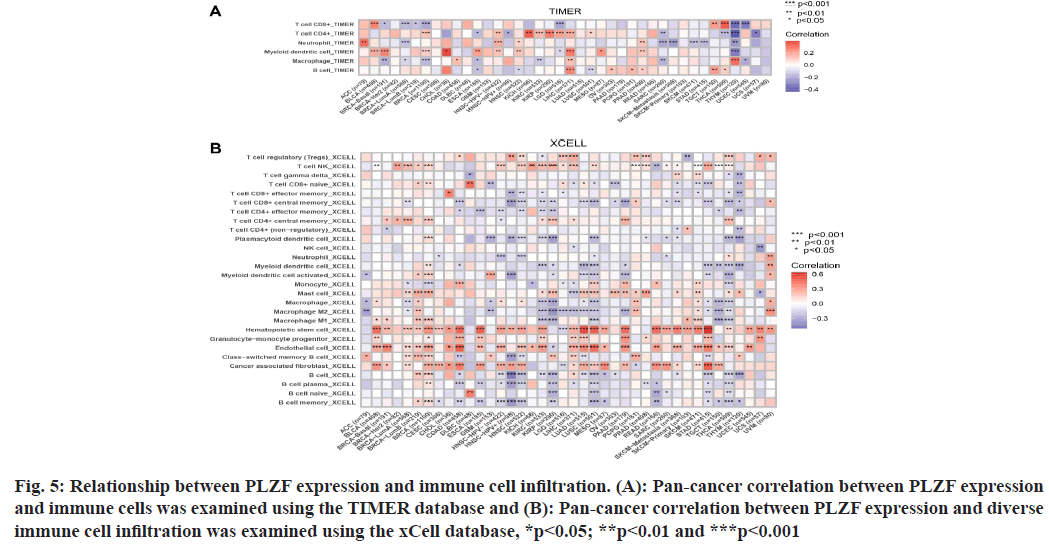
Fig. 5: Relationship between PLZF expression and immune cell infiltration. (A): Pan-cancer correlation between PLZF expression and immune cells was examined using the TIMER database and (B): Pan-cancer correlation between PLZF expression and diverse immune cell infiltration was examined using the xCell database, *p<0.05; **p<0.01 and ***p<0.001
Because the distinct relationship between PLZF and immunity, we performed a pan-cancer analysis of the association between PLZF expression and immune cell infiltration based on data from the TIMER database. As shown in fig. 6A, expression of PLZF was significantly associated with an abundance of the following tumorinfiltrating immune cells; CD8+ T cells in 10 types of cancer, CD4+ T cells in 14 types of cancer, neutrophils in 10 types of cancer, macrophages in 12 types of cancer and B cells in 9 types of cancer.
We further used the cell online tool to examine the relationship between PLZF expression and the infiltration of different immune cell subtypes. Among the 28 subtypes of immune cells, we found that PLZF expression was significantly correlated with immune cell infiltration in BLCA, BRCA, COAD, LUAD, LUSC, PAAD, READ and STAD. Hematopoietic stem cells, endothelial cells and cancer-associated fibroblasts were most positively associated with PLZF expression in these different cancers (fig. 6B).
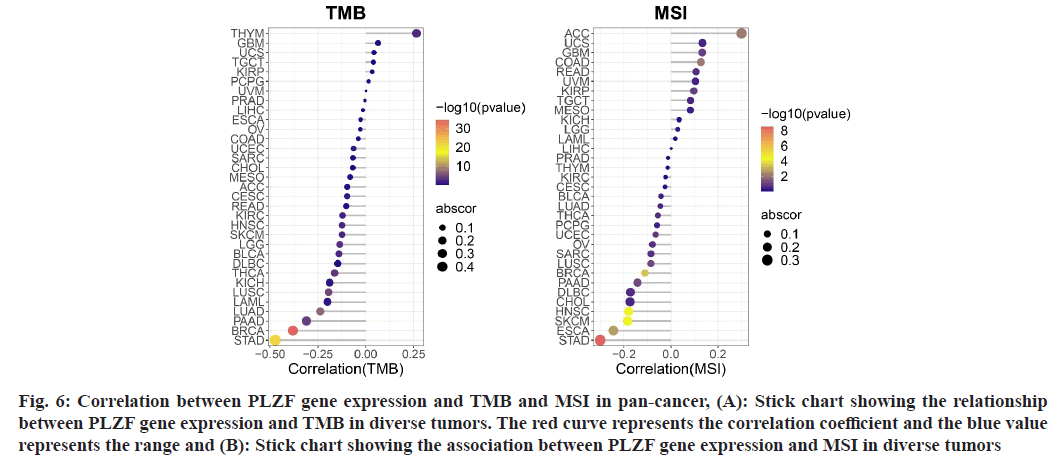
Fig. 6: Correlation between PLZF gene expression and TMB and MSI in pan-cancer, (A): Stick chart showing the relationship between PLZF gene expression and TMB in diverse tumors. The red curve represents the correlation coefficient and the blue value represents the range and (B): Stick chart showing the association between PLZF gene expression and MSI in diverse tumors
Because TMB and MSI are two emerging biomarkers believed to be associated with response to immunotherapy, we went on to examine the relationship between PLZF expression and TMB[13-15]. Notably, expression of PLZF correlated with TMB in several cancer types including THYM, KIRC, HNSC, LGG, BLCA, THCA, LUSC, LUAD, PAAD and BRCA (fig. 7A). We also investigated the correlation between PLZF expressions with MSI in 33 different types of cancer. Positive correlations between PLZF expression and MSI were found in ACC and COAD, while negative correlations were observed among cases of BRCA, HNSC, SKCM, ESCA and STAD (fig. 7B).
Fig. 7: PLZF-related gene enrichment analysis, (A): Identification of experimentally determined PLZF-binding proteins using
the STRING tool; (B): Analysis of the correlation between the expression of PLZF and selected target genes, including KLF9,
TSC22D3, SUN2, KLF15, and TNS2. The top 100 PLZF-correlated genes were identified using the GEPIA2 approach; (C):
Corresponding heat map data for various cancer types; (D): Intersection analysis of PLZF-binding proteins and products of genes
with correlated expression profiles; (E): KEGG pathway analysis based on PLZF-binding proteins and related genes and (F): Biological
process, cellular component and molecular function data from GO analysis
Note: ( ): BP; (
): BP; ( ): CC and (
): CC and ( ): MF
): MF
PLZF is a DNA-binding protein with sequence specificity which is mainly localized in the nucleus and widely expressed in the brain, lung and endocrine tissues with some degree of sex-dependence. Previous studies have linked the expression of PLZF with growth, development and effector functions, indicating that PLZF may potentially be a reliable diagnostic and prognostic marker for several human cancer types[16-19]. However, no pan-cancer study of PLZF has been carried out to date. In addition, the molecular mechanisms behind how PLZF may affect the pathogenesis of various cancers remain to be explored. To explore this topic, we summarized the features of gene expression and genetic alterations of PLZF across 33 different cancer types using data from multiple databases, including TCGA, CPTAC, KEGG and Gene Expression Omnibus (GEO).
Our analysis found that the expression of PLZF was significantly decreased in BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LUAD, LUSC, READ, STAD and UCEC samples relative to tumor-adjacent tissues. In recent years, studies have confirmed that the expression of PLZF is significantly decreased in gastric, lung, pancreatic and prostate cancers[9,20]. Thus, decreasing PLZF has been suggested to inhibit cell migration and tumor growth, which is consistent with our observations.
Kaplan-Meier analysis revealed that PLZF expression was correlated with OS prognosis in OV, STAD, KICH, KIRC and LIHC. Though the high expression of PLZF has been shown to correlate with good OS for Non- Small Cell Lung Cancer (NSCLC) patients, including for patients with LUAD[21]. We found that decreased expression of PLZF was correlated with worse DFS for patients with KICH, KIRC, KIRP and THCA. Decreased expression of PLZF generally predicted poor OS and DFS, which suggested that PLZF may be a useful prognostic biomarker for cancer patients.
KEGG and GO analyses revealed potential molecular mechanisms linking PLZF to cancer. We found that several genes including KLF9, TSC22D3, SUN2, KLF15 and TNS2 had PLZF-related expression profiles. Our enrichment analysis demonstrated that PLZF may exert its tumorigenic effects by protein binding, metal ion binding, chromatin binding, identical protein binding and transcription factor binding.
Previous experiments have demonstrated that low pH extracellular environments can suppress Natural Killer T (NKT) cell function by inhibiting the nuclear translocation of PLZF and mammalian Target of Rapamycin (mTOR) signaling[22]. As NKT cells possess anti-tumor activity, they are good candidate for targets of immunotherapy and clinical drug trials that aim to modulate their activity, which are currently under way[23]. This suggests that PLZF may be a promising potential target for future anticancer immunotherapies. Another study suggested that lactate in the tumor microenvironment reduces Peroxisome Proliferator- Activated Receptor Gamma (PPARγ) expression in invariant NKT (iNKT) cells within the tumor, thereby reducing lipid biogenesis and Interferon-γ (IFN-γ) synthesis[24]. Production of IFN-γ is a key function of iNKT cells, so PPARγ and PLZF may enhance anti-tumor immunity by synergizing with iNKT cell activation to increase lipid synthesis. PLZF is therefore of great importance in iNKT cell-based anti-cancer immunotherapy. However, few studies exist that explore the immune function and anticancer effects of PLZF.
As either a tumor suppressor or oncogene, PLZF plays an important role in the occurrence and development of various tumors. Clinically, PLZF assays can aid in the early diagnosis of cancers, assessment of staging and grading, selection of optimal treatment methods, and prediction of surgical prognosis. Furthermore, PLZF represents a potential therapeutic target that may be harnessed as part of iNKT cell-based antitumor immunotherapy. However, the immune response to cancer and its modulation by physiological and pharmacological factors are complex multilevel processes which are still relatively poorly understood. Therefore, there is an urgent need for in vivo and in vitro experiments exploring this area in greater depth to aid in the development of new anticancer therapies.
Funding:
This work was supported by Huaian Science and Technology Development Fund (grant number: HAB201924).
Author’s contributions:
Jiru Wang and Yuan Zhang have contributed equally to this work and were considered as first authors.
Conflict of interests:
The authors declared no conflict of interests.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Yoshimaru T, Nakamura Y, Katagiri T. Functional genomics for breast cancer drug target discovery. J Hum Genet 2021;66(9):927-35.
[Crossref] [Google Scholar] [PubMed]
- Malki A, ElRuz RA, Gupta I, Allouch A, Vranic S, Al Moustafa AE. Molecular mechanisms of colon cancer progression and metastasis: Recent insights and advancements. Int J Mol Sci 2020;22(1):130.
[Crossref] [Google Scholar] [PubMed]
- Bergholz JS, Wang Q, Kabraji S, Zhao JJ. Integrating immunotherapy and targeted therapy in cancer treatment: Mechanistic insights and clinical implications. Clin Cancer Res 2020;26(21):5557-66.
[Crossref] [Google Scholar] [PubMed]
- Spicuglia S, Vincent-Fabert C, Benoukraf T, Tibéri G, Saurin AJ, Zacarias-Cabeza J, et al. Characterisation of genome-wide PLZF/RARA target genes. PLoS One 2011;6(9):e24176.
[Crossref] [Google Scholar] [PubMed]
- Licht JD, Chomienne C, Goy A, Chen A, Scott AA, Head DR, et al. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17). Blood 1995;85(4):1083-94.
[Crossref] [Google Scholar] [PubMed]
- Shen H, Zhan M, Zhang Y, Huang S, Xu S, Huang X, et al. PLZF inhibits proliferation and metastasis of gallbladder cancer by regulating IFIT2. Cell Death Dis 2018;9(2):1-4.
[Crossref] [Google Scholar] [PubMed]
- Hsieh CL, Botta G, Gao S, Li T, van Allen EM, Treacy DJ, et al. PLZF, a tumor suppressor genetically lost in metastatic castration-resistant prostate cancer, is a mediator of resistance to androgen deprivation therapy. Cancer Res 2015;75(10):1944-8.
[Crossref] [Google Scholar] [PubMed]
- Wang JB, Jin Y, Wu P, Liu Y, Zhao WJ, Chen JF, et al. Tumor suppressor PLZF regulated by lncRNA ANRIL suppresses proliferation and epithelial mesenchymal transformation of gastric cancer cells. Oncol Rep 2019;41(2):1007-18.
[Crossref] [Google Scholar] [PubMed]
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47(W1):W556-60.
[Crossref] [Google Scholar] [PubMed]
- Genova C, Dellepiane C, Carrega P, Sommariva S, Ferlazzo G, Pronzato P, et al. Therapeutic implications of tumor microenvironment in lung cancer: Focus on immune checkpoint blockade. Front Immunol 2021;12:799455.
[Crossref] [Google Scholar] [PubMed]
- Lu Y, Li Y, Liu Q, Tian N, Du P, Zhu F, et al. MondoA–thioredoxin-interacting protein axis maintains regulatory t-cell identity and function in colorectal cancer microenvironment. Gastroenterology 2021;161(2):575-91.
[Crossref] [Google Scholar] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 2019;51(2):202-6.
[Crossref] [Google Scholar] [PubMed]
- Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22(11):1342-50.
[Crossref] [Google Scholar] [PubMed]
- Yang G, Zheng RY, Jin ZS. Correlations between microsatellite instability and the biological behaviour of tumours. J Cancer Res Clin Oncol 2019;145(12):2891-9.
[Crossref] [Google Scholar] [PubMed]
- Hu S, Chen Y, Liu L, Yin X, Yang Y, Tang L. PLZF and PLZF-MAPK10 can predict the prognosis of postoperative patients with hepatocellular carcinoma. Int J Clin Exp Pathol 2020;13(12):3158.
[Google Scholar] [PubMed]
- Han H, Wang S, Meng J, Lyu G, Ding G, Hu Y, et al. Long noncoding RNA PART1 restrains aggressive gastric cancer through the epigenetic silencing of PDGFB via the PLZF-mediated recruitment of EZH2. Oncogene 2020;39(42):6513-28.
[Crossref] [Google Scholar] [PubMed]
- Zhang Q, Li X, Li Y, Chen S, Shen X, Dong X, et al. Expression of the PTEN/FOXO3a/PLZF signalling pathway in pancreatic cancer and its significance in tumourigenesis and progression. Invest New Drugs 2020;38(2):321-8.
[Crossref] [Google Scholar] [PubMed]
- Jin Y, Nenseth HZ, Saatcioglu F. Role of PLZF as a tumor suppressor in prostate cancer. Oncotarget 2017;8(41):71317.
[Crossref] [Google Scholar] [PubMed]
- Xiao GQ, Li F, Findeis-Hosey J, Hyrien O, Unger PD, Xiao L, et al. Down-regulation of cytoplasmic PLZF correlates with high tumor grade and tumor aggression in non–small cell lung carcinoma. Hum Pathol 2015;46(11):1607-15.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Wang L, Guo S, Bao Y, Ma Y, Yan F, et al. Hypermethylation reduces expression of tumor-suppressor PLZF and regulates proliferation and apoptosis in non-small-cell lung cancers. FASEB J 2013;27(10):4194-203.
[Crossref] [Google Scholar] [PubMed]
- Prevot N, Pyaram K, Bischoff E, Sen JM, Powell JD, Chang CH. Mammalian target of rapamycin complex 2 regulates invariant NKT cell development and function independent of promyelocytic leukemia zinc-finger. J Immunol 2015;194(1):223-30.
[Crossref] [Google Scholar] [PubMed]
- Payne KK, Bear HD, Manjili MH. Adoptive cellular therapy of cancer: Exploring innate and adaptive cellular crosstalk to improve anti-tumor efficacy. Future Oncol 2014;10(10):1779-94.
[Crossref] [Google Scholar] [PubMed]
- Fu S, He K, Tian C, Sun H, Zhu C, Bai S, et al. Impaired lipid biosynthesis hinders anti-tumor efficacy of intratumoral iNKT cells. Nat Commun 2020;11(1):1-5.
[Crossref] [Google Scholar] [PubMed]