- *Corresponding Author:
- Hui Cai
The First Clinical Medical College of Lanzhou University, Lanzhou, Gansu 730000,China
E-mail: caialonteam@163.com
| This article was originally published in a special issue, “Drug Development in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(5) Spl Issue “129-143” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
The immune costimulatory protein V-set domain containing T cell activation inhibitor 1 is a key molecule in the B7 family developing advanced growth in multiple cancers as a suppressive regulatory factor during T cell immunoreaction. For the purpose of confirming the importance of prognosis and ascertaining the effects on tumor immunity of V-set domain containing T cell activation inhibitor 1 gene expression across diverse cancers. The design utilized a wide range of tools involved in bioinformatics from the available public database to integrally identify the association between V-set domain containing T cell activation inhibitor 1 and prognosis, clinic pathological features, tumor mutational burden, microsatellite instability, methyl transferase, mismatch repair, immune cell permeation among different tumors, chemotherapy drug sensitivities, and try to comprehensively probe potentially carcinogenic of V-set domain containing T cell activation inhibitor 1. The results exhibited that V-set domain containing T cell activation inhibitor 1 was overexpressed in various tumor types. As an independent risk factor for the prognosis of 12 different cancer types, V-set domain containing T cell activation inhibitor 1 expression had a connection with survival status. Additionally, it is advantageous in 15 different cancer types for diagnosis. Meanwhile, in 10 and 7 cancer types, the expression of V-set domain containing T cell activation inhibitor 1 was distinctly correlated to tumor mutational burden and microsatellite instability. And the close connection of V-set domain containing T cell activation inhibitor 1 with immune cell infiltration was considered, especially T cells cluster of differentiation 8+, T cells cluster of differentiation 4+, neutrophils, myeloid dendritic cells, macrophages and B cells within most types of cancer. The implication of V-set domain containing T cell activation inhibitor 1 showed the valuable prognostic potentiality in multiple cancers, and it may function as an immunotherapy target in multiple malignant tumors according to the impact on tumorigenesis and tumor immunity.
Keywords
V-set domain containing T cell activation inhibitor 1, pan-cancer, prognosis, tumor immune cell infiltration, tumor microenvironment
Based on present ranks and recent trends, cancer is the material cause of premature death this century across 57 countries, including China[1]. The burden of cancer incidence and mortality worldwide is growing rapidly and the types of cancer are enormously diverse[2]. Correspondingly, cancer has become a burdensome obstacle to lengthening life expectancy in every country in the world[2]. With the continuous progress of medical technology, in recent years, immunotherapy has been deemed an essential instrument to improve the prognosis of cancer treatment for numerous suffers with multiple hematological and solid malignancies[3]. Evidently, the therapeutic schedule of immune checkpoint blockade has led to a vital pattern in the fight against cancer[4]. Immune checkpoint inhibitors target the dysfunctional immune system to induce cancer-cell killing by Cluster of Differentiation (CD) 8-positive T cells[5]. Recently, the immunologic checkpoint suppressants (anti-Cytotoxic-T-Lymphocyte-Associated Antigen 4 (CTLA-4) and anti-Programmed Cell Death (PD) protein antibodies are represented) have proved remarkable long-term anticancer efficacy in various cancers, and their clinical success highlighted the central role of the immune system in cancer control and provided a new strategy for cancer treatment[6].
V-Set Domain Containing T Cell Activation Inhibitor 1 (VTCN1), also referred to as B7x, B7S1, or B7-H4, was initially identified by bioinformatics determination from three independent laboratories, 24, 37-39, and later demonstrated the existence of the expression of VTCN1 messenger Ribonucleic Acid (mRNA) and VTCN1 protein in human serous ovarian and breast cancers[7]. VTCN1, as a B7 family member was authenticated in 2003[8]. B7 family was constituted by a series of associated co-stimulators in a structure which can combine the receptors of lymphocytes to adjust the immunoreaction[9]. Relevant studies have shown that VTCN1 negative regulation of the immune response and inhibits the T cells proliferation, cytokine secretion, and cell cycle involved to promote immune escape[10]. VTCN1 contains a Nuclear Localization Sequence (NLS) motif making it function as a cytoplasmic-nuclear shuttle protein[11]. Certainly, VTCN1 has become a major participant in the genesis and developed progress of tumors, such as cell multiplication, anti-apoptosis, invasion, metastasis, etc., which is connected to the poor outcome and high recurrence rate in tumor patients[8]. There was plenty of studies that have suggested that VTCN1 can be employed in tumor diagnosis as a biomarker due to constitutively expressed in various tumors[10]. Furthermore, VTCN1 is a significant molecular intervention target for the treatment of cancer[12]. Overall, a variety of experiments have indicated that VTCN1 is strongly correlated with tumor, inflammation, autoimmune diseases along with organ transplantation.
Antibody drugs, like anti-PD-1 and anti-PD-Ligand 1 (PD-L1), substantiated worthy contributions including widespread feasibility which is not limited to the special case and the durability of clinical reaction during an efficacious plan. Nevertheless, the total response rates remain dissatisfactory, as far as cancers with low mutational burden are concerned especially[4]. For another, up to now, major research investigating the VTCN1 function in tumors has been restricted to only certain categories. No study based on generic cancer has been done on the relationship between VTCN1 and various cancers.
Accordingly, further exploration of VTCN1 and interference with this negative regulatory is essential and profound. Therefore, in this study, we comprehensively used multifarious data storage systems which included the Cancer Genome Atlas (TCGA), the Cancer Cell Line Encyclopedia (CCLE), Genotype-Tissue Expression (GTEx), and cBioPortal to explore the merits of prognosis and ascertaining the effects on tumor immunity of VTCN1 gene expression across diverse cancers.
Materials and Methods
Differential VTCN1 expression in pan-cancer:
The VTCN1 data collection about 33 kinds of cancer and relevant clinical material was delivered from TCGA by virtue of the UCSC cancer genome browser (https://xenabrowser.net/datapages/). The VTCN1 expression data obtained by TCGA and GTEx formed an expression matrix to process. The tumor and adjacent normal tissues of distinguishing expression about VTCN1 was evaluated. In addition, the expression of VTCN1 in different cell lines of 32 cancer types was obtained from the CCLE (https://portals.broadinstitute.org/ccle/about).
VTCN1 of survival, Receiver Operating Characteristic (ROC) curve analysis and clinic pathology in pan-cancer:
Survival-related data were gathered for each cancer which was downloaded through the TCGA. For the implication of VTCN1 and prognosis, the Overall Survival (OS), Disease-Specific Survival (DSS), Disease-Free Interval (DFI), and Progression-Free Interval (PFI) were chosen to carry on a systematic comprehension and analyzed the correlation between VTCN1 expression and the above indexes of the 33 cancers. Taking the intermediate value of VTCN1 expression level as a standard, the patient's division was into two groups, high and low. Kaplan-Meier method was confronted with prognostic outcomes between these two groups. The “survival” and “forestplot” in R packages were used for achieving forest plots. The area under the ROC curves (Area Under the Curve (AUC)) was calculated to determine the diagnostic value. The correlational analyses of clinic pathology were primarily executed using the “limma” and “ggpubr” of the R-package.
Association with Tumor mutational burden (TMB), Microsatellite Instability (MSI), methyltransferase and Mismatch Repair (MMR) in pan-cancer:
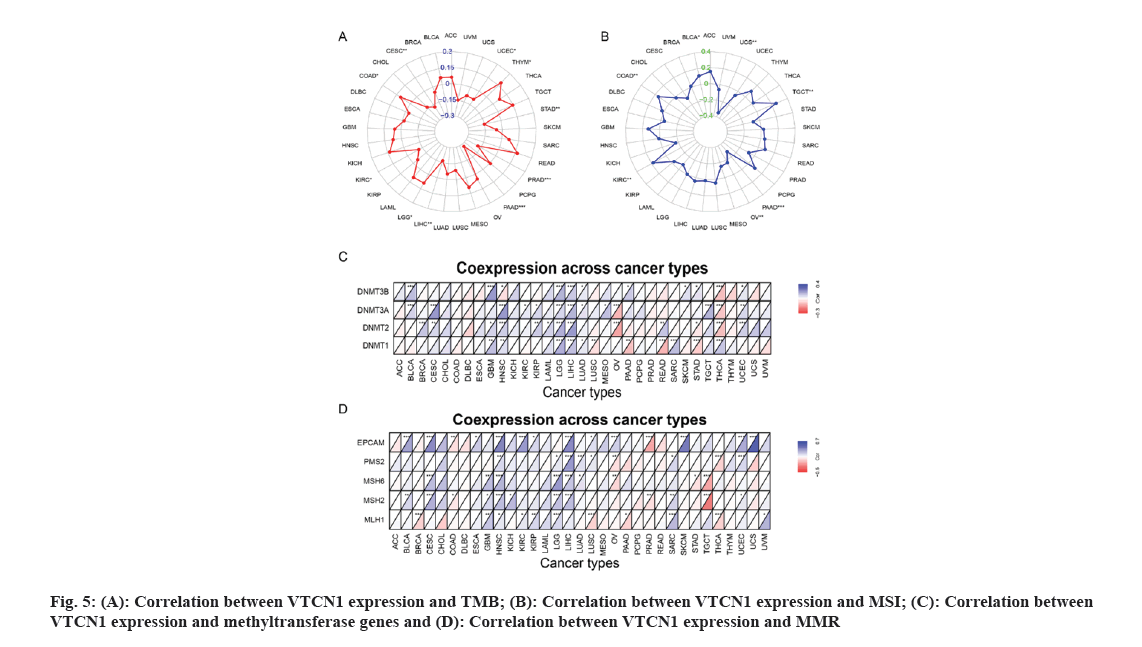
The MSI scores were derived from TCGA. TMB represent a certain number of base mutations per million bases to determine the frequency of mutations in the different tumor samples. Spearman's method to perform both correlation analyses for TMB and MSI. Both metrics were visualized by a radar map. The methyltransferase and MMR gene expression levels were assessed using TCGA expression profile data. Using the R packages “reshape2” and “RColorBrewer”, the output was shown as a heat map.
Relationship between VTCN1 and immunity in pan-cancer:
We established an evaluation of stromal and immune cells in tumor tissues utilizing (ESTIMATE) to intuitively compute correspondent scores. Then the correlation analysis for these 2 scores proceeded accordingly. Results of the relationship between VTCN1 expression and immune checkpoint co-expression using the R package functional output. Meanwhile, the Tumor Immune Estimation Resource (TIMER) database was used to analyze the infiltration of immune cells in tumor tissue. We further estimated the immune cell infiltration in 33 kinds of cancers was performed by MCPCOUNTER, QUANTISEQ and CIBERSORT to obtain a computation of the abundance of specialized cell types.
Genes and pathways associated with the immunology of VTCN1:
From the GSEA website (https://www.gsea-msigdb.org/gsea/downloads.jsp), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) datasets were downloaded. Both GO functional annotation and KEGG signal path enrichment of VTCN1 were delineated by the R-packages “limma” “org. Hs. eg. db” “cluster Profiler” and “enrichplot”.
Drug sensitivity analysis of VTCN1 in pan-cancer:
The drug sensitivity analysis of VTCN1 in pan-cancer was conducted using NCI-60 composition data and gene-expression arrays from the CallMiner™ (https://discover.nci.nih. gov/cellminer/home.do). Clinical trial or FDA-approved medications were considered. The “impute”, “limma”, “ggplot2”, and “ggpubr” R package were performed.
Cell culture, RNA isolation and Reverse Transcription-Polymerase Chain Reaction (RT-PCR):
SW620 and SW480 colorectal adenocarcinoma cells, MCF-7 breast cancer cells, CAKI kidney cancer cells were placed in a humidified 5 % Carbon dioxide (CO2) incubator at 37° and cultured in RPMI-1640 containing 10 % Fetal Bovine Serum (FBS) (Invitrogen), 1 % double antibodies (streptomycin and penicillin) (Corning), while NCM460 normal colonic epithelial cells, MCF-10A normal mammary epithelial cells and HK2 normal renal tubular epithelial cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) medium under the same conditions. RNA was extracted as required by the M5 Universal RNA Mini Kit, and to determine RNA concentration and purity compliance, absorbance values were measured at 260 nm and 280 nm. RNA was reverse transcribed to cDNA according to the instructions of the M5 Sprint qPCR RT kit with gDNA remover reverse transcription kit. 2× M5 HiPer SYBR Premix EsTaq (with Tli RNaseH) was used as a fluorescent dye for RT-qPCR assay. VTCN1 primers were designed and synthesized by Wuhan Servicebio Technology Co. The sequences were as follows: forward, 5'- TCTCGGAAGTCTCCAATACCAGC-3'; reverse, 5'- TCTCGGAAGTCTCCAATACCAGC -3'. RT-qPCR reaction conditions were as follows; 95° for 30 s; 95° for 5 s, 60° for 30 s, 40 cycles; 60° for 20 s, 95° for 20 s. VTCN1 mRNA expression levels were calculated and analyzed by the 2-ΔΔCt formula.
Data management:
Data-processing was consolidated transformation using log2. The Wilcox test was conducted on two sample types; the p<0.05 was statistical significance. The Kaplan-Meier curve and Cox proportional hazard regression model were applied for all survival analyses. The Spearman’s rank method to discuss the correlation between two variables; p<0.05 were identified as significant. The R software made data visualization processing (***p< 0.001; **p<0.01 and **p<0.05).
Results and Discussion
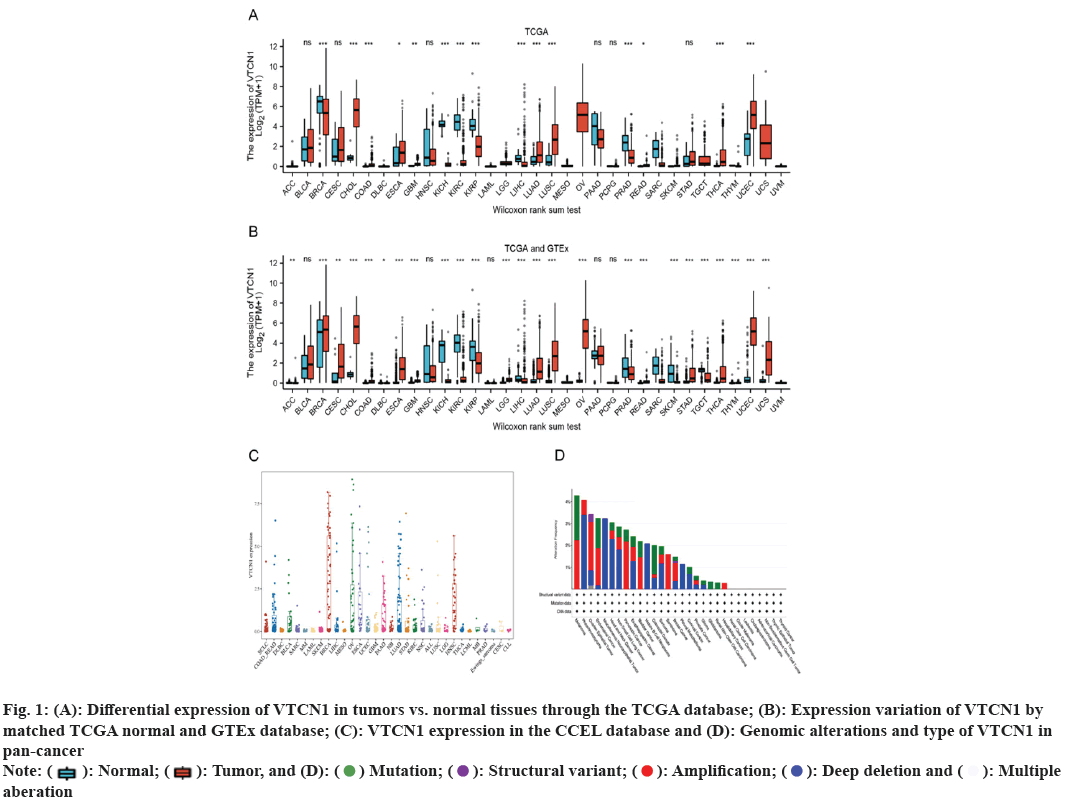
Primitively, in addition to normal tissue data being absent, differences in VTCN1 expression between each group were discovered in 15 kinds of cancer. Among them, VTCN1 was the high expression in 9 tumor types including Cholangiocarcinoma (CHOL), Colon Adenocarcinoma (COAD), Esophageal Carcinoma (ESCA), Glioblastoma Multiforme (GBM), Lung Adenocarcinoma (LUAD), Lung Squamous Cell Carcinoma (LUSC), Rectum Adenocarcinoma (READ), Thyroid Carcinoma (THCA) and Uterine Corpus Endometrial Carcinoma (UCEC). Conversely, VTCN1 levels were low expressed in Breast Invasive Carcinoma (BRCA), Kidney Chromophobe (KICH), Kidney Renal Clear Cell Carcinoma (KIRC), Kidney Renal Papillary Cell Carcinoma (KIRP), Liver Hepatocellular Carcinoma (LIHC), and Prostate Adenocarcinoma (PRAD). It was remarkable that the most striking expressive distinction in tumors compared to normal tissues was kidney cancer. In addition, several cancers just had a very small number of normal samples, like sarcoma (SARC), and these differences could not be determined to be statistically significant (fig. 1A). Meanwhile, matching TCGA normal and GTEx database, VTCN1 was highly expressed in Adrenocortical Carcinoma (ACC), cervical squamous cell Carcinoma and Endocervical Adenocarcinoma (CESC), brain Lower Grade Glioma (LGG), Ovarian Serous Cystadenocarcinoma (OV), Stomach Adenocarcinoma (STAD) and Thymoma (THYM), lowly expressed in lymphoid neoplasm Diffuse Large B-cell lymphoma (DLBC), Skin Cutaneous Melanoma (SKCM) and Tenosynovial Giant Cell Tumor (TGCT), compared to normal tissue (fig. 1B). Additionally, through the violin diagram obtained in the CCEL database, VTCN1 was highly expressed in six cancers: BRCA, OV, ESCA, Pancreatic Adenocarcinoma (PAAD), LUAD and Head and Neck Squamous Cell Carcinoma (HNSC) (fig. 1C). To explore the genomic alterations of VTCN1 gene levels across all cancers, we presently evaluated the alteration frequency situation for each cancer. In 22 cancers, VTCN1 had mutation or copy number variations. As a result, in melanoma, endometrial cancer, and colorectal cancer, VTCN1 mutated highly (fig. 1D).
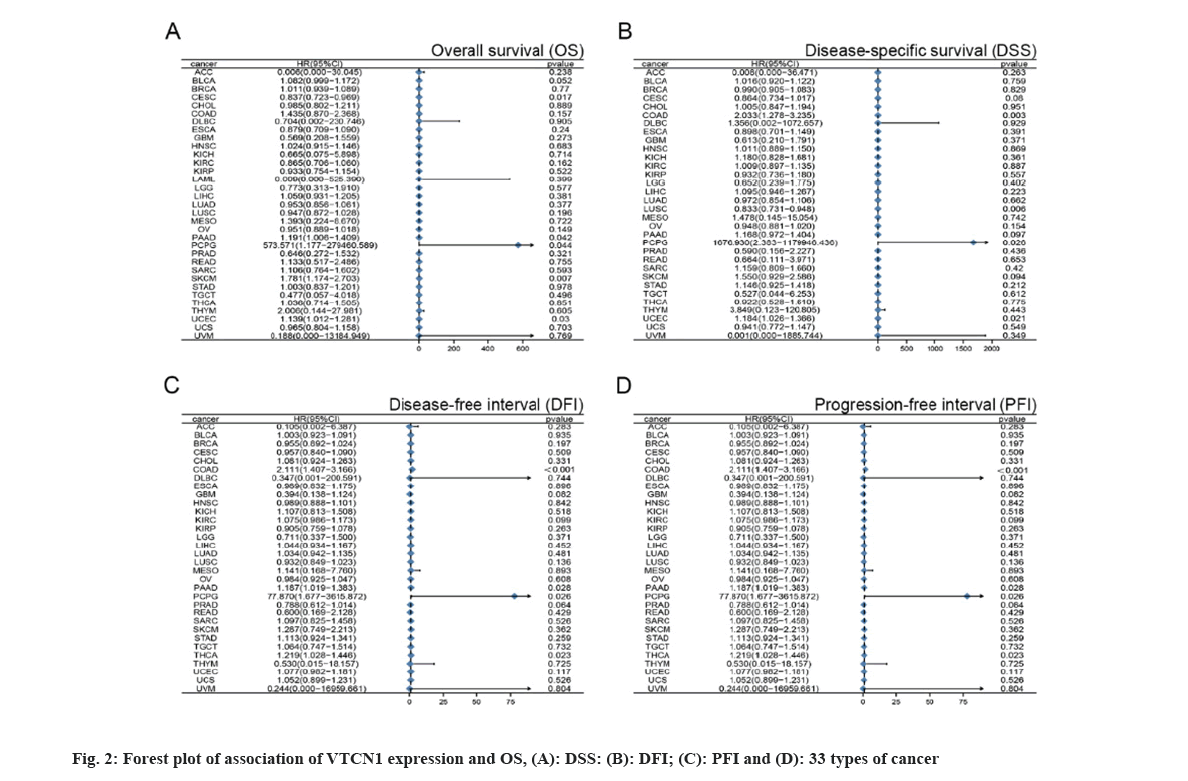
With the purpose of figuring out the prognostic discrepancies affected by VTCN1 expression level, we conducted a correlativity test by prognostic measures in all cancers that included OS, DSS, DFI, and PFI. First, VTCN1 expression was linked to OS, DSS, DFI, and PFI in patients with each of the four malignancies, according to the forest plots produced by the statistical approach of Student-test. (fig. 2) Overall, the prognosis of COAD, Pheochromocytoma and Paraganglioma (PCPG), PAAD, THCA and UCEC patients was most significantly correlated with VTCN1 expression.
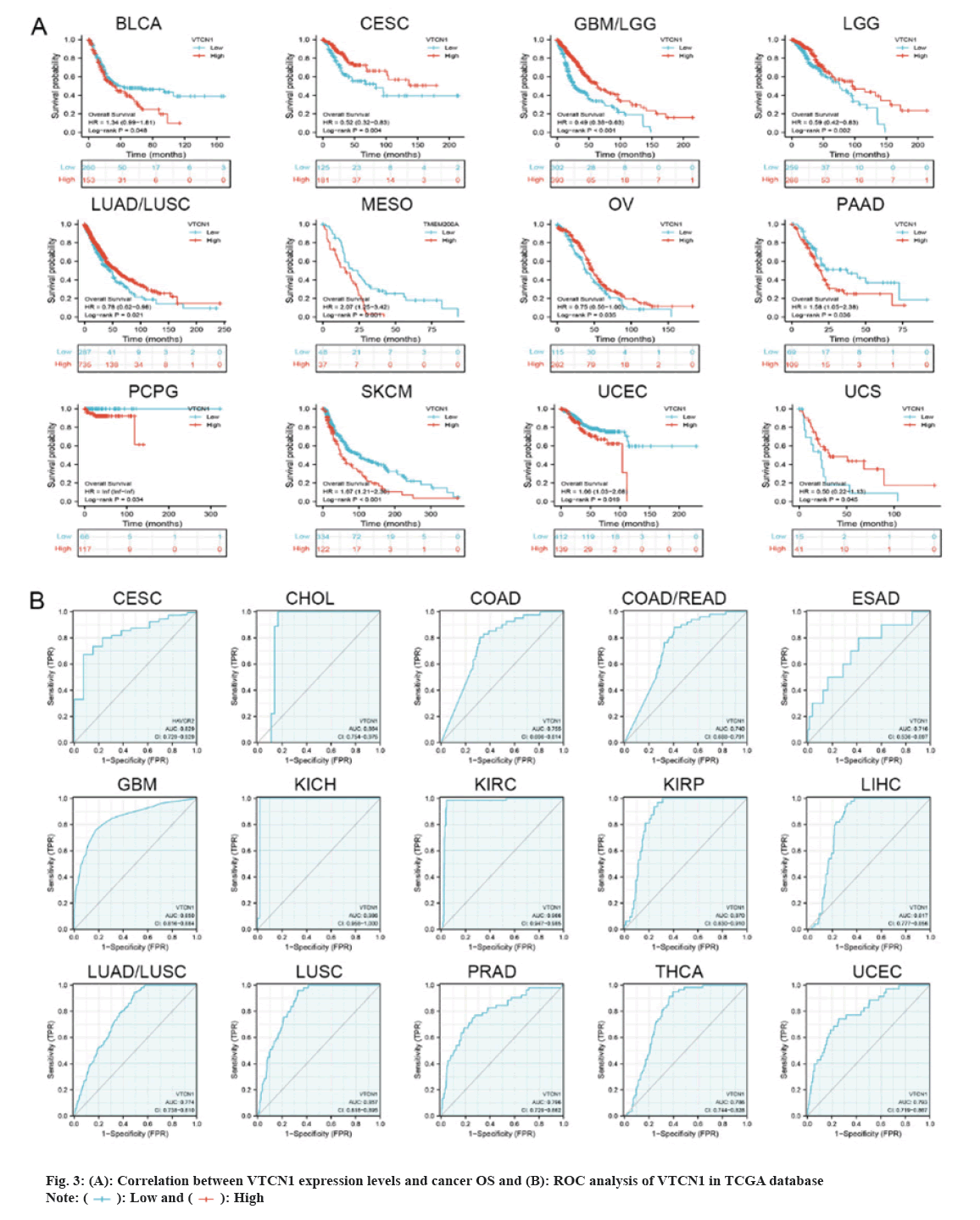
Then, we determined that VTCN1 expression levels were a risk factor for OS in patients with 12 malignancies using COX regression analysis. From the Kaplan-Meier curves of OS, we could conclude that increased VTCN1 expression exhibited a poor prognosis in Bladder Urothelial Carcinoma (BLCA), Mesothelioma (MESO), PAAD, PCPG, SKCM and UCEC. Diametrically, patients in CESC, GBM/LGG, LGG, LUAD/LUSC, OV and Uterine Carcinosarcoma (UCS) with higher VTCN1 expression benefited from significantly longer survival rates compared to those with lower expression as shown in fig. 3A.
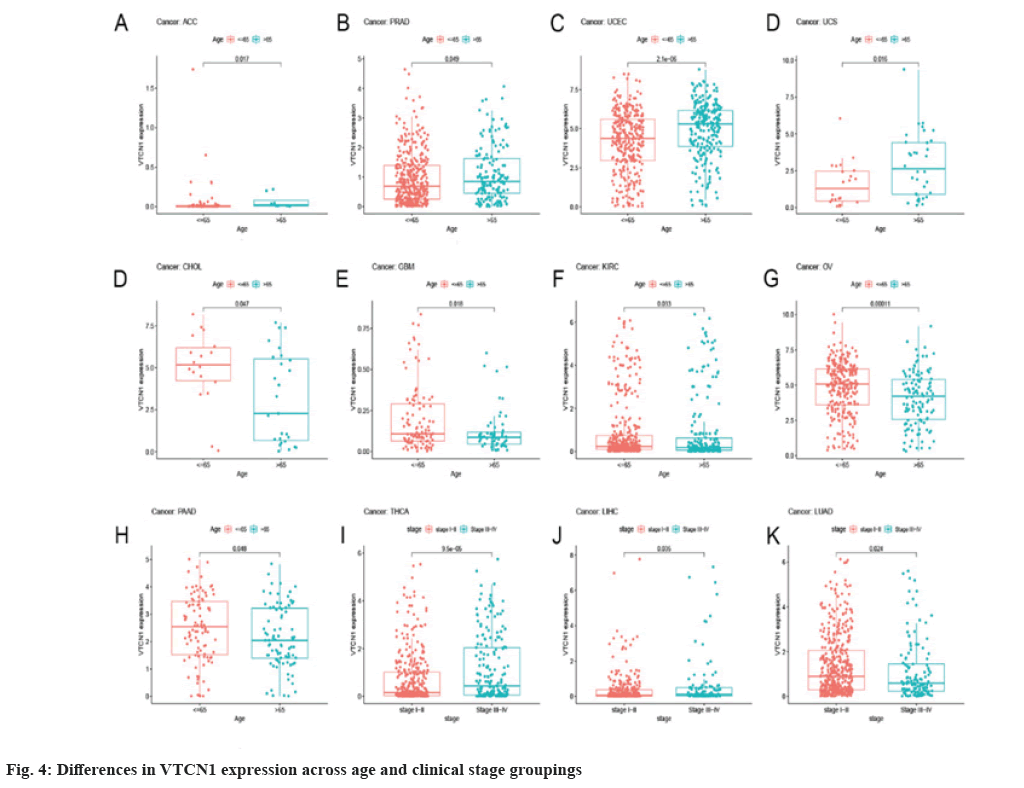
The Receiver Operating Characteristic (ROC) curve has shown diagnostic function for a variety of tumors. VTCN1 has moderate diagnostic accuracy for 15 types of cancer, especially for LUSC, KIRP, KIRC, KICH, GBM and CHOL has higher diagnostic value as shown in fig. 3B.Furthermore, as for the discovery of the relationship between VTCN1 expression and clinic pathological phenotypes, we compared the expression differences of VTCN1 according to two patient age stratification in every cancer. Specifically, we observed that those older than 65 y with ACC, PRAD, UCEC, and UCS had higher expression levels, however, regarding CHOL, GBM, KIRC, OV and PAAD, higher VTCN1 expression was found in patients younger than 65 y old. No pronounced correlations between age and VTCN1 levels were demonstrated in other cancers. Correspondingly, VTCN1 expression was disparate in several tumors because of the different clinical stages by analysis. VTCN1 had a high level in stage I-II patients but was infrequent in stage III-IV in THCA. As for LIHC and LUAD, VTCN1 was overexpressed in stage III-IV, differing from stage I-II patients as shown in fig. 4.
TMB has been of use as a biomarker across many cancer types for diagnosis and can be served as an information provider about immune checkpoint blockade therapy efficacy which may benefit patients undergoing immunotherapy[13]. Therefore, detecting the relationship between TMB and VTCN1 expression in various cancers is essential. Our data demonstrated that TMB has a significant correlation with VTCN1 expression in several specific cancers. There, VTCN1 had a positive association with TMB in three types, COAD, LGG, and THYM inclusive. Interestingly, VTCN1 had an inverse association with TMB in other seven types, including PAAD, PRAD, CESC, LIHC, STAD, KIRC, and UCEC as shown in fig. 5A.
MSI came to play an important part in the evaluation of malignancy, efficacy and prognosis of the tumor[14]. In this connection, we ulteriorly confirmed an association between VTCN1 expression and MSI in diverse forms of cancer. The data certified that there was an association between VTCN1 expression and MSI in 7 cancer types. Respectively, these four cancers had an upward correlation with MSI: COAD, KIRC, TGCT and BLCA. On the contrary, a discernible negative relationship was discovered between VTCN1 and MSI in the other three cancer types, including PAAD, UCS and OV as shown in fig. 5B.
We next investigated more into the relationships between VTCN1 expression, methyltransferase and MMR. Our analysis of the heat map results revealed a positive correlation between VTCN1 expression and methyltransferase, particularly in GBM, HNSC, LGG and LIHC (fig. 5C). Similarly, in CESC, HNSC, LGG and LIHC, the expression of genes involved in MMR was directly related to VTCN1 expression as shown in fig. 5D.
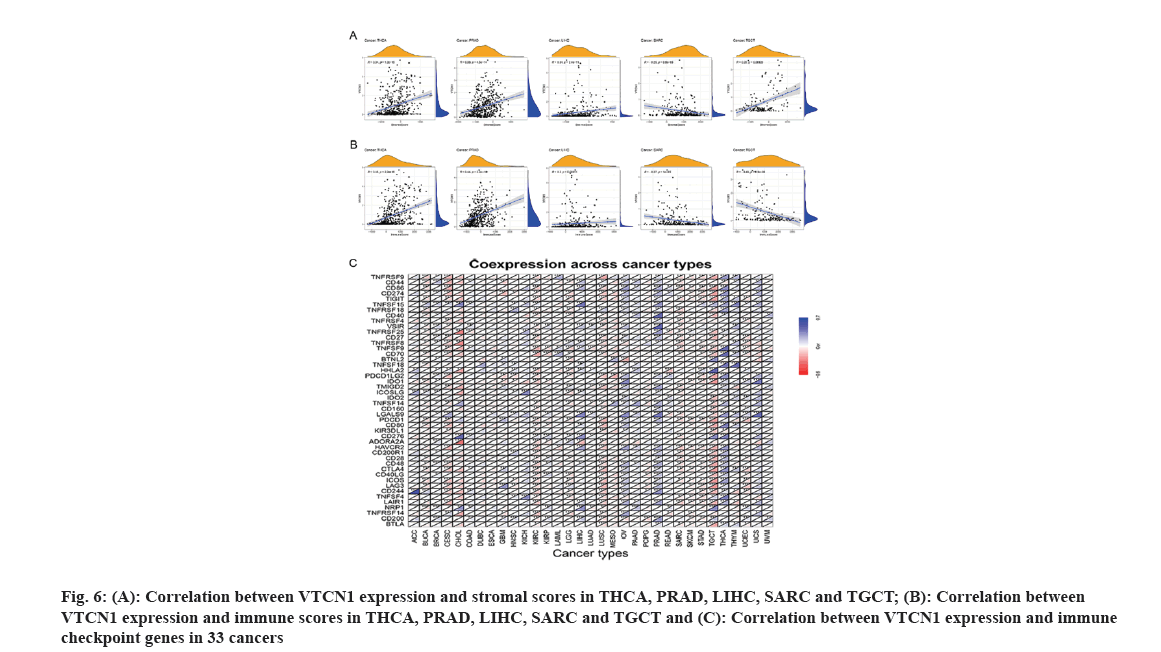
It is well-known that TME takes a pivotal part during tumor initiation, progression and metastasis. Simultaneously, the persistent crosstalk between tumor cells and their surrounding stromal creates environment-mediated drug resistance. Hence, it is profound to further elucidate the contact characteristics between TME and VTCN1 expression for enlarging our understanding of the pathological mechanism in pan-cancer. For a clear quantification of the immune and matrix components in cancer, an organic calculation about stromal and immune cell scores in cancers has been made. The findings showed that a positive correlation of VTCN1 expression with both stromal and immune scores in THCA, PRAD and LIHC, while the scores of stromal and immune in SARC had a negative relationship with VTCN1 expression. Interestingly, we noticed that VTCN1 expression was positively related to stromal score but negatively with an immune score in TGCT (fig. 6A and fig. 6B). In fact, VTCN1 has been shown to be positively correlated with a number of immune checkpoint genes that have been thoroughly investigated in various malignancies, which offered compelling evidence that VTCN1 might serve as a new therapeutic target as shown in fig. 6C.
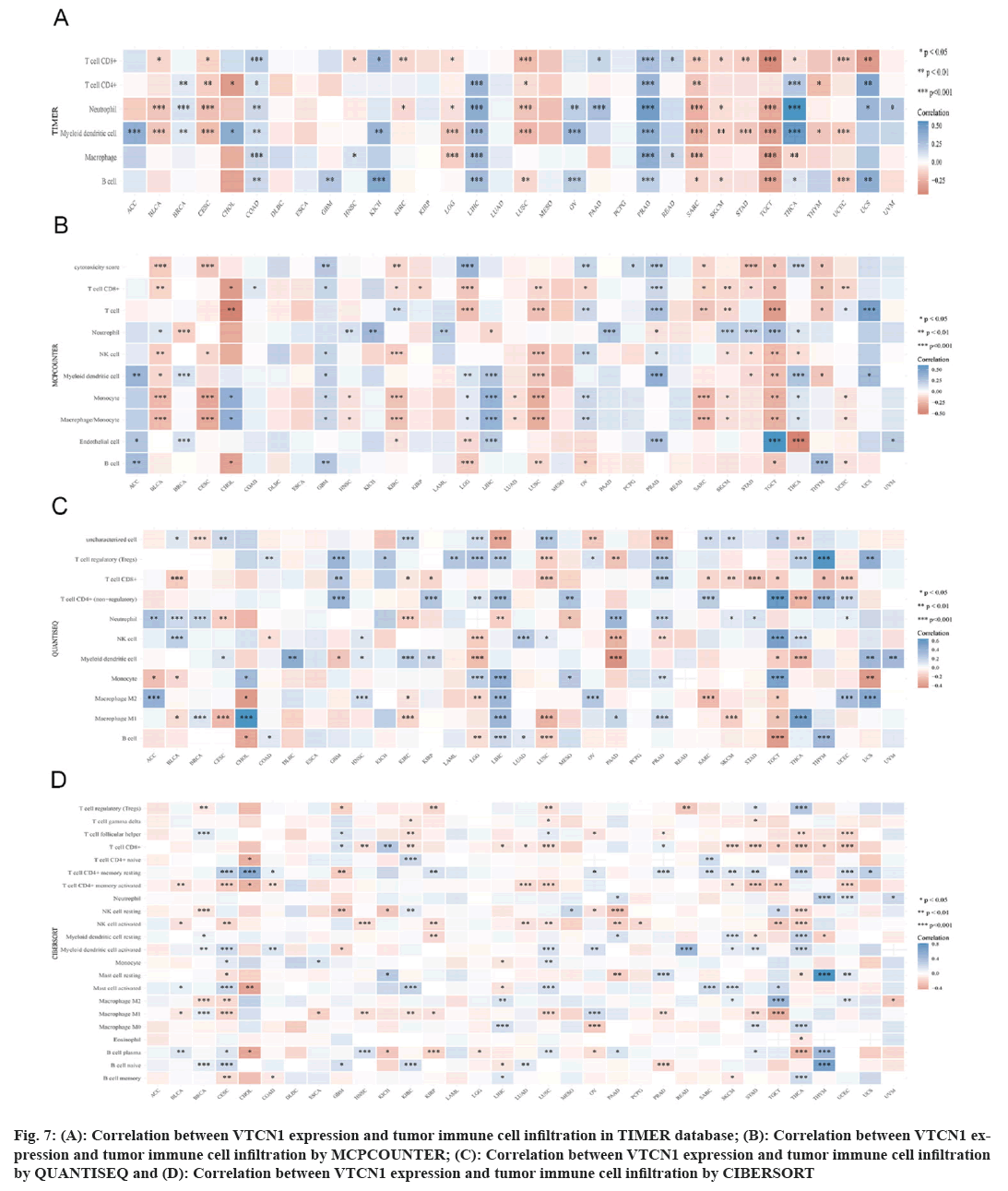
Immune cells can both support or hinder therapeutic efficacy, mutually their activated state and localization can vary under the influence of the TME[15]. In this case, we used four different algorithms, the results indicated that the prevalence of tumor immune cell infiltration had an obvious correlation with VTCN1 in most, especially in LIHC, PRAD, SARC, TGCT, and THCA. VTCN1 expression submitted a distinct pertinence with multifarious infiltrating immune cells, like T cells CD8+, T cells CD4+ memory resting, NK cell, myeloid dendritic cell activated, mast cell activated, B cell plasma, and macrophages as shown in fig. 7.
Fig 7: (A): Correlation between VTCN1 expression and tumor immune cell infiltration in TIMER database; (B): Correlation between VTCN1 expression and tumor immune cell infiltration by MCPCOUNTER; (C): Correlation between VTCN1 expression and tumor immune cell infiltration by QUANTISEQ and (D): Correlation between VTCN1 expression and tumor immune cell infiltration by CIBERSORT
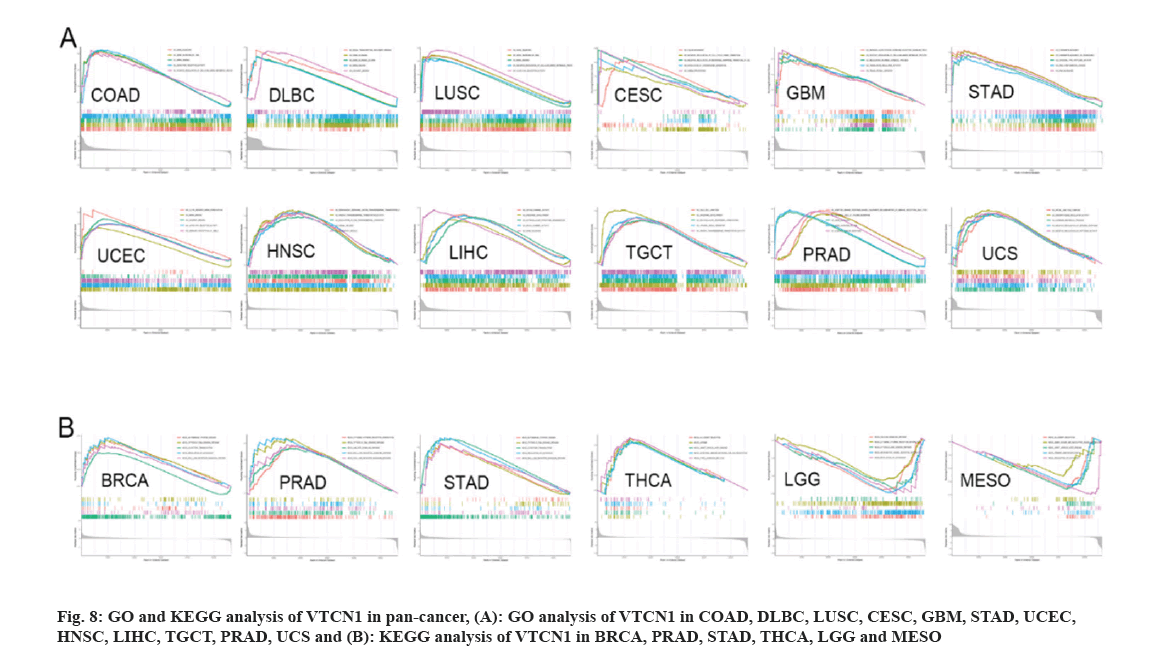
Pan-cancer of GO functional annotation and KEGG pathway sets was conducted to verify the biological characteristics of VTCN1. In the condition of GO, VTCN1 is a positive regulator to participate in essential biology involving gene silencing, gene silencing by RNA, mRNA binding, etc. across COAD, DLBC, and LUSC. For another, VTCN1 actively regulated and intimately provided several functions related to biological activities in different types of cancer. These activities included; gene expression processes, including processing, shear in transcription and translation and cell proliferation (such as regulation of the cell cycle), differentiation in CESC, GBM, STAD, and UCEC; and signaling pathways on cell surface receptors in HNSC, LIHC, and TGCT; regulation of immune responses in PRAD and UCS (fig. 8A). The analysis of KEGG enrichment indicated that VTCN1 expression was concerned with the cytosolic DNA sensing pathway and several pivotal immune cell-related pathways involving, T cell receptor signaling pathway; intestinal immune network for IgA production; cytokine receptor interaction; JAK-STAT signaling pathway; Rig I like receptor signaling pathways; Toll-like receptor signaling pathway; regulation of autophagy and allograft rejection in BRCA, PRAD, STAD, and THCA. Contrarily, in LGG and MESO, VTCN1 negatively regulated several specific immune-related pathways as shown in fig. 8B.
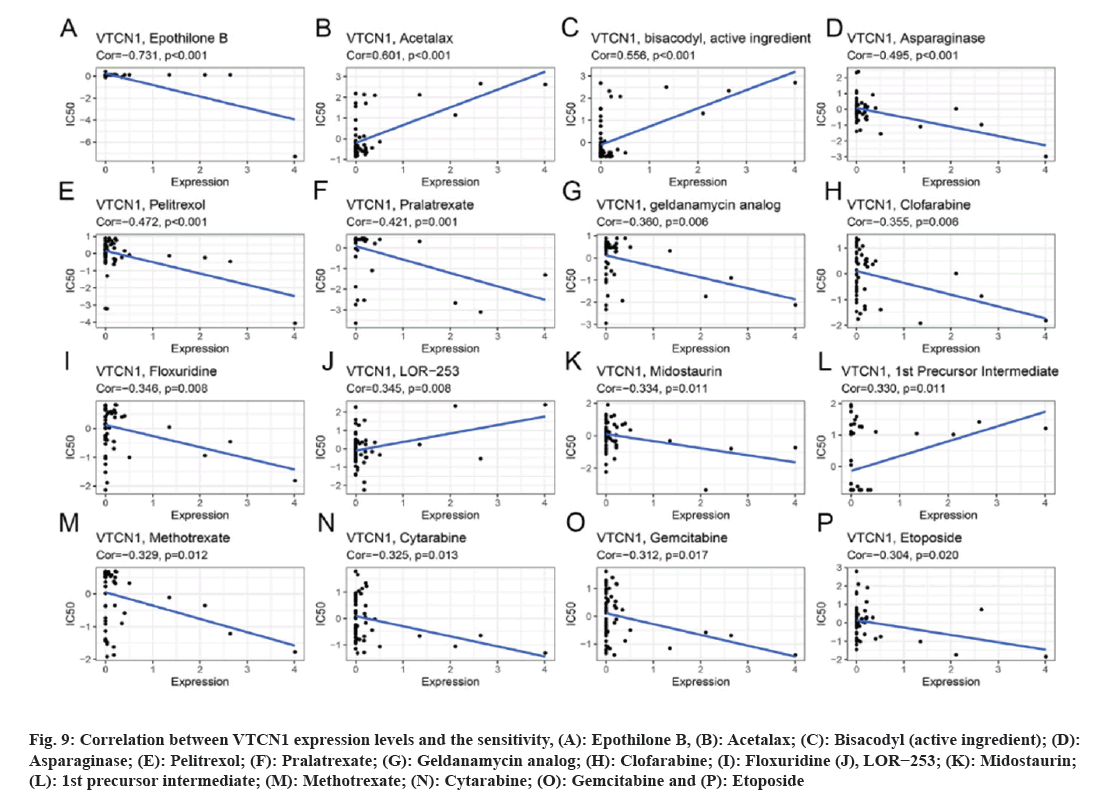
Data results for our application of CellMiner™ database to analyze the potential link between VTCN1 expression and drug sensitivity are indicated the VTCN1 expression levels had a positive relation with acetalax, bisacodyl (active ingredient), LOR−253 and 1st precursor intermediate sensitivity (fig. 9A-fig. 9C, fig. 9J and fig. 9L). Conversely, VTCN1 expression was negatively correlated with drug susceptibility of epothilone B, asparaginase, pelitrexol, pralatrexate, geldanamycin analog, clofarabine, floxuridine, midostaurin, methotrexate, cytarabine, gemcitabine, etoposide (fig. 9A, fig. 9D-fig. 9P). According to the findings, VTCN1 expression was linked to treatment resistance to some routinely prescribed clinical anticancer medications, including gemcitabine and etoposide.
Fig 9: Correlation between VTCN1 expression levels and the sensitivity, (A): Epothilone B, (B): Acetalax; (C): Bisacodyl (active ingredient); (D): Asparaginase; (E): Pelitrexol; (F): Pralatrexate; (G): Geldanamycin analog; (H): Clofarabine; (I): Floxuridine (J), LOR−253; (K): Midostaurin; (L): 1st precursor intermediate; (M): Methotrexate; (N): Cytarabine; (O): Gemcitabine and (P): Etoposide
Experimental verification:
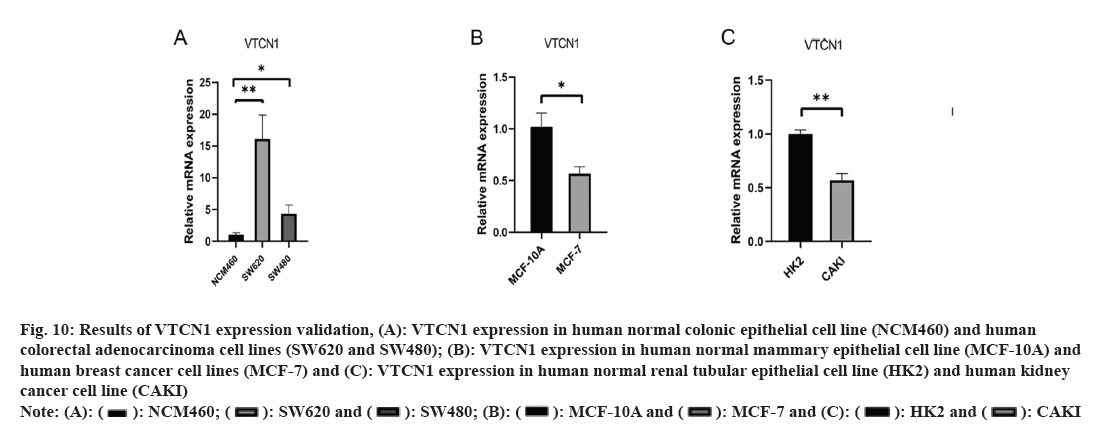
We applied RT-PCR to reach a deeper understanding of VTCN1 mRNA expression. The experimental results indicated that VTCN1 expression was higher in colorectal cancer, simultaneously, lower in breast cancer and kidney cancer, which was consistent with the bioinformatics analysis as shown in fig. 10.
Fig 10: Results of VTCN1 expression validation, (A): VTCN1 expression in human normal colonic epithelial cell line (NCM460) and human colorectal adenocarcinoma cell lines (SW620 and SW480); (B): VTCN1 expression in human normal mammary epithelial cell line (MCF-10A) and human breast cancer cell lines (MCF-7) and (C): VTCN1 expression in human normal renal tubular epithelial cell line (HK2) and human kidney cancer cell line (CAKI) 
Notably, based on B7H4 immunotherapy formulations consisting of mono-cloned antibodies, ADC, anti-B7-H4/CD3BsAb, and CAR-T cells have had an original creation and a generalized therapeutic effect was achieved[10]. We excavated the similarities and variations of VTCN1 in tumors to contribute to a better understanding and insight into immune target therapy. Our research findings were consistent with previous reports published, in which VTCN1 was overexpressed in CHOL[16], COAD[17], GBM[18], LUAD[19], LUSC[19], THCA[20], UCEC[21], ESCA[22], OV[21] and Oral Squamous Cell Carcinoma (OSCC)[23]. There are relevant studies to confirm the mRNA levels of VTCN1 in Thymic Epithelial Tumor exhibiting remarkable associativity with the World Health Organization pathological classification[24]. Nevertheless, when comparing the TCGA databases, some contradiction about VTCN1 expression variation were observed. For example, our data showed low expression of VTCN1 in LIHC, contradicting Dong's and colleague’s report[25], while the research has found that VTCN1 expression was no significant difference between the liver tumor tissues and normal tissues[26]. These discrepancies were due to the differences in tumor samples and special approaches to data collection as well as potential differences in the biological mechanisms involved. The possibility of VTCN1 having research significance is supported by its differential expression in tumor tissues as opposed to normal tissues.
From the comprehensive statistics of all data of our survival analysis, we found that high levels of VTCN1 predicted poor survival in PAAD, COAD, SKCM and BLCA patients, but high levels of VTCN1 expression are prognostically beneficial in patients with LGG. In Shen and colleagues' report, VTCN1 was also considered a factor of poor prognosis in pancreatic cancer[27]. Ding and his colleagues reported that VTCN1 overexpression was associated with a poor prognosis in colorectal cancer patients[28]. Mizuno et al. through Cox multivariate regression analysis of 133 patients with upper urothelial carcinoma who had the operation of nephroureterectomy demonstrated that higher VTCN1 expression was a noted predictor for shorter OS[29]. Correspondingly, an independent study found that patients with VTCN1 low expressing melanoma benefited from longer survival rates by Quandt et al., which is consistent with our results[30]. One study confirmed that B7-H4 activation on Mφs/microglia in the microenvironment of gliomas is an important immunosuppressive event blocking effective T-cell immune responses which leads to poor prognosis in glioma patients[18]. Additionally, it was well reported that VTCN1 was an independent risk factor for the prognosis of ovarian cancer[31]. A targeted approach to treatment may be possible given that the level of VTCN1 expression correlates with the prognosis of many malignancies. According to our research, the expression of VTCN1 was also crucial for the detection of various cancers.
Furthermore, there is an increasing body of literature that demonstrates that the VTCN1 expression levels in various cancers correlate with tumor stage[8]. Moreover, we observed that VTCN1 was age-related in 9 types of cancers. VTCN1 expression was interrelated with the clinical stage in THCA, LIHC and LUAD was also revealed in our study. Our original discovery can provide a reference value for subsequent treatment.
Certainly, TMB may serve as a predictor for clinical response to ICI during the preceding reporter and observe the intermediate association of higher TMB and survival rate improvement[32]. The majority of research has indicated, with higher MSI levels could achieve more competent anti-tumor immune advantages, inhibit tumor cell growth better, and improve prognosis preferably relative to those with low MSI levels or who are microsatellite stable 14. In our study, an evident association of VTCN1 expression with TMB and MSI in 10 and 7 cancer types respectively. We speculated that the process of VTCN1 expression resulted in a rise in tumor burden and ultimately tumorigenesis in CESC, HNSC, LGG, and LIHC when combined with our analysis of the findings of the correlation between VTCN1 expression and methylation transferase and MMR respectively.
It is now clear that the immune cells within the TME take a considerable part in tumorigenesis[6]. The bulk of the researchers have reported the mechanism of the mutual effect between VTCN1 and infiltrating immune cells leading to tumorigenesis. Overexpression of VTCN1 accelerates renal cell carcinoma advancement by recruitment of tumor-related neutrophils up-regulating the CXCL8 in the report by Li et al[33]. Others included cervical cancer[34], thymic epithelialtumor[24], epithelial ovarian cancer[35], metastatic colorectal cancer[28], and non-small cell lung cancer[36], where the connection between VTCN1 and immune cells was explained severally. Correspondingly, our exploration aimed at VTCN1 connection with the tumor microenvironment and infiltrating immune cells also found the existence of distinct associations in the majority of cancers, especially with activated dendritic cells, macrophages, T cells CD4+, T cells CD8+ and mast cells resting. It was known that VTCN1, through inhibiting the immune responses of effector T-cells realizes immune evasion. Cheng with partners detected that the secretion of Interleukin (IL)-10, Tumor Nucrosis Factor Alpha (TNF-α), and Interferon-Gamma (IFN-γ) by T cells were raised after blocking VTCN1 expression on tumor infiltration of DCs[37], and Treg cells increased the secretion levels of IL-10 by APCs, subsequently generating APC VTCN1 expression and presentation of these APCs immunosuppressive[38]. Additionally, we discovered that the expression of VTCN1 was favorably linked with a number of immune checkpoint inhibitors in various malignancies. Furthermore, VTCN1 has been correlated with resistance to a few commonly used chemotherapeutic medications. As a result, VTCN1 may integrate seamlessly with other cancer-treating medications. Immunoglobulin G (IgG) 1B7-H4 is the ongoing exploitation for cancer treatment and manifests a dramatic safety profile in IND-enabling research[39]. XMT-1660, an antibody-drug conjugate in the composition of an anti-B7-H4 antibody conferring great link-coupled specificity. It has been confirmed consistently showed more significant anti-tumor ability in the model within the organism in breast cancer[40]. Thus, immunosuppressive agents targeting VTCN1 will benefit more patients. Collectively, based on these findings and our research, the clinical therapeutic potential and significance of targeting VTCN1 are ponderable. The tumor microenvironment could be favorably altered within the therapeutic blockade of VTCN1, allowing for antigen-specific clearance of tumor cells.
Additionally, enrichment analyses exhibited the ability of VTCN1 to affect cancer etiology or pathogenesis by acting on gene expression and immunomodulatory pathways. Our data can be supported by previously published articles. The experiment proved that VTCN1 acts on carcinogenesis and cell proliferation via translocation into the nucleus as a cytoplasmic-nuclear shuttling protein[11,41]. Actually, research has shown that the Phosphoinositide 3-Kinase (PI3K)/Protein Kinase B (AKT) signaling pathway might inhibit the nuclear translocation of VTCN1[42]. The protein is highly expressed on tumors but low expressed on normal tissue makes VTCN1 an attractive directly targeted molecule with low off-target toxicity. Regrettably, the receptor for VTCN1 has not yet been determined, further exploration is still needed.
Our research revealed that VTCN1 expression is significantly associated with colorectal cancer and contributes to tumorigenesis, diagnosis, prognosis and immunological processes. The first study, by Sadun et al., established the link between the tumor escape mechanism in colorectal cancer and a notable uptick in the expression of the immunosuppressive gene VTCN1[43]. Recently, the study found that VTCN1 promoted colorectal cancer cell proliferation, migration, and invasion by triggering the epithelial-mesenchymal transition[44]. These findings demonstrate the great potential research value of VTCN1 in colorectal cancer and motivate us to further explore its role.
Despite our first study of the role of VTCN1 in pan-cancer, there are some limitations. First, the research mainly revolved around the biological information of VTCN1 expression and patient clinical and pathological features via the application of various online tools that lacked more powerful verification of experimental tests in vivo and in vitro. Second, a systematic bias may arise due to the variability in database establishment and specimen collection, and some possible mechanisms with disruptive biological characteristics between each cancer. The specific role of VTCN1 in different types of cancer needs forward investigation using it as a springboard. Looking into the future, we will continue to focus on prospective studies of VTCN1 in different cancers, as well as clinical drug trial results. On the basis of this study, we are going to explore the specific mechanism at the cellular or molecular level.
The emphasis is on the potential value of VTCN1 during the development and progression of tumors by means of synthetic bioinformatics methods in this study. As a prospective cancer therapeutic target, the distinguishing expression exists in tumors and normal tissues and the affection on the prognosis of pan-cancer patients. Besides, VTCN1 expression was related to clinic pathological features and the TMB, MSI, methyltransferase, MMR, some chemotherapy drug sensitivities, as well as immune cell content in various cancer types. Therefore, worthwhile opinion presence and a strong theoretical cornerstone were offered for making progress in alternative immune modulatory therapies and improving the efficacy of cancer immunotherapy.
Acknowledgment:
The authors thank the Key Laboratory of Molecular Diagnostics and Precision Medicine for Surgical Oncology in Gansu Province and the da Vinci Surgery System Database (DSSD, www.davincisurgerydatabase.com) for their help and support in the methodology process.
Funding:
This work was funded by the 2021 Central-Guided Local Science and Technology Development Fund (ZYYDDFFZZJ-1), Major projects of Joint Scientific research Fund (23JRRA1537), National Natural Science Foundation of China (23JRRA1320), Gansu daVinci robot high-end diagnosis and treatment team construction project, Key Research and Development Plan of Gansu Province (No. 21YF5FA169), Natural Science Foundation of Gansu Province (No. 22JR11RA257, No. 22JR5RA692, No. 21JR7RA633, No. 21JR1RA038), Research project of Traditional Chinese Medicine of Gansu Province (GZKZ-2022-6), and Gansu Province Excellent Doctor Fund Project (23JRRA1320).
Author contributors:
Jingyao Ren and Yongfeng Wang conceived and designed the study and revised the manuscript. Jingyao Ren, Yongfeng Wang, Bangqian Mo, Xianglai Jiang, and Lihui Zhu conducted all data collection and analysis, and compiled charts. All authors read and approved the final manuscript.
Conflict of interests:
The authors declared no conflict of interests.
References
- Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021;127(16):3029-30.
[Crossref] [Google Scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Kruger S, Ilmer M, Kobold S, Cadilha BL, Endres S, Ormanns S, et al. Advances in cancer immunotherapy 2019–latest trends. J Exp Clin Cancer Res 2019;38(1):268.
[Crossref] [Google Scholar] [PubMed]
- He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res 2020;30(8):660-9.
[Crossref] [Google Scholar] [PubMed]
- Carlino MS, Larkin J, Long GV. Immune checkpoint inhibitors in melanoma. Lancet 2021;398(10304):1002-14.
[Crossref] [Google Scholar] [PubMed]
- Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett 2020;470:126-33.
[Crossref] [Google Scholar] [PubMed]
- Podojil JR, Miller SD. Potential targeting of B7-H4 for the treatment of cancer. Immunol Rev 2017;276(1):40-51.
[Crossref] [Google Scholar] [PubMed]
- Wang L, Heng X, Lu Y, Cai Z, Yi Q, Che F. Could B7-H4 serve as a target to activate anti-cancer immunity? Int Immunopharmacol 2016;38:97-103.
[Crossref] [Google Scholar] [PubMed]
- Yu N, Li X, Zheng S, Li X. B7-H4’s role “beyond the tumor”. Inflammation 2013;36(4):941-7.
[Crossref] [Google Scholar] [PubMed]
- Wang JY, Wang WP. B7-H4, a promising target for immunotherapy. Cell Immunol 2020;347:104008.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, Wu H, Lu D, Li G, Sun C, Song H, et al. The costimulatory molecule B7-H4 promote tumor progression and cell proliferation through translocating into nucleus. Oncogene 2013;32(46):5347-58.
[Crossref] [Google Scholar] [PubMed]
- MacGregor HL, Ohashi PS. Molecular pathways: Evaluating the potential for B7-H4 as an immunoregulatory target. Clin Cancer Res 2017;23(12):2934-41.
[Crossref] [Google Scholar] [PubMed]
- Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol 2019;30(1):44-56.
[Crossref] [Google Scholar] [PubMed]
- Yang G, Zheng RY, Jin ZS. Correlations between microsatellite instability and the biological behaviour of tumours. J Cancer Res Clin Oncol 2019;145(12):2891-9.
[Crossref] [Google Scholar] [PubMed]
- Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett 2017;387:61-8.
[Crossref] [Google Scholar] [PubMed]
- Xie N, Cai JB, Zhang L, Zhang PF, Shen YH, Yang X, et al. Upregulation of B7-H4 promotes tumor progression of intrahepatic cholangiocarcinoma. Cell Death Dis 2017;8(12):3205.
[Crossref] [Google Scholar] [PubMed]
- Peng HX, Wu WQ, Yang DM, Jing R, Li J, Zhou FL, et al. Role of B7-H4 siRNA in proliferation, migration and invasion of LOVO colorectal carcinoma cell line. Biomed Res Int 2015;2015:326981.
[Crossref] [Google Scholar] [PubMed]
- Yao Y, Ye H, Qi Z, Mo L, Yue Q, Baral A, et al. B7-H4 (B7x)–mediated cross-talk between glioma-initiating cells and macrophages via the IL6/JAK/STAT3 pathway lead to poor prognosis in glioma patients. Clin Cancer Res 2016;22(11):2778-90.
[Crossref] [Google Scholar] [PubMed]
- Sun Y, Wang Y, Zhao J, Gu M, Giscombe R, Lefvert AK, et al. B7-H3 and B7-H4 expression in non-small-cell lung cancer. Lung Cancer 2006;53(2):143-51.
[Crossref] [Google Scholar] [PubMed]
- Zhu J, Chu BF, Yang YP, Zhang SL, Zhuang M, Lu WJ, et al. B7-H4 expression is associated with cancer progression and predicts patient survival in human thyroid cancer. Asian Pac J Cancer Prev 2013;14(5):3011-5.
[Crossref] [Google Scholar] [PubMed]
- Tringler B, Liu W, Corral L, Torkko KC, Enomoto T, Davidson S, et al. B7-H4 overexpression in ovarian tumors. Gynecol Oncol 2006;100(1):44-52.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Wang L, Wang W, Zhao L, Shan B. B7-H4 facilitates proliferation of esophageal squamous cell carcinoma cells through promoting interleukin-6/signal transducer and activator of transcription 3 pathway activation. Cancer Sci 2016;107(7):944-54.
[Crossref] [Google Scholar] [PubMed]
- Wu L, Deng WW, Yu GT, Mao L, Bu LL, Ma SR, et al. B7-H4 expression indicates poor prognosis of oral squamous cell carcinoma. Cancer Immunol Immunother 2016;65(9):1035-45.
[Crossref] [Google Scholar] [PubMed]
- Yan X, Feng J, Hong B, Qian Y. The expression of PD-L1 and B7-H4 in Thymic epithelial tumor and its relationship with tumor immune-infiltrating cells. Front Oncol 2021;11:662010.
[Crossref] [Google Scholar] [PubMed]
- Dong L, Xie L, Li M, Dai H, Wang X, Wang P, et al. Downregulation of B7-H4 suppresses tumor progression of hepatocellular carcinoma. Sci Rep 2019;9(1):14854.
- Qian Y, Shen L, Cheng L, Wu Z, Yao H. B7-H4 expression in various tumors determined using a novel developed monoclonal antibody. Clin Exp Med 2011;11:163-70.
[Crossref] [Google Scholar] [PubMed]
- Shen L, Qian Y, Wu W, Weng T, Wang FX, Hong B, et al. B7-H4 is a prognostic biomarker for poor survival in patients with pancreatic cancer. Hum Pathol 2017;66:79-85.
[Crossref] [Google Scholar] [PubMed]
- Ding S, Lv X, Liu Z, Zhan S, Xu Y, Zhang X, et al. Overexpression of B7-H4 is associated with infiltrating immune cells and poor prognosis in metastatic colorectal cancer. Int Immunopharmacol 2021;90:107144.
[Crossref] [Google Scholar] [PubMed]
- Mizuno T, Kamai T, Tsuzuki T, Nishihara D, Kijima T, Arai K, et al. Elevated expression of B7 homolog 4 is associated with disease progression in upper urinary tract urothelial carcinoma. Cancer Immunol Immunother 2022;71(3):565-78.
[Crossref] [Google Scholar] [PubMed]
- Quandt D, Fiedler E, Boettcher D, Marsch WC, Seliger B. B7-H4 expression in human melanoma: Its association with patient’s survival and antitumor immune response. Clin Cancer Res 2011;17(10):3100-11.
[Crossref] [Google Scholar] [PubMed]
- Xu M, Zhang B, Zhang M, Liu Y, Yin FL, Liu X, et al. Clinical relevance of expression of B7-H1 and B7-H4 in ovarian cancer. Oncol Lett 2016;11(4):2815-9.
[Crossref] [Google Scholar] [PubMed]
- Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genetics 2019;51(2):202-6.
[Crossref] [Google Scholar] [PubMed]
- Li A, Zhang N, Zhao Z, Chen Y, Zhang L. Overexpression of B7-H4 promotes renal cell carcinoma progression by recruiting tumor-associated neutrophils via upregulation of CXCL8. Oncol Lett 2020;20(2):1535-44.
[Crossref] [Google Scholar] [PubMed]
- Han S, Wang Y, Shi X, Zong L, Liu L, Zhang J, et al. Negative roles of B7-H3 and B7-H4 in the microenvironment of cervical cancer. Exp Cell Res 2018;371(1):222-30.
[Crossref] [Google Scholar] [PubMed]
- MacGregor HL, Garcia-Batres C, Sayad A, Elia A, Berman HK, Toker A, et al. Tumor cell expression of B7-H4 correlates with higher frequencies of tumor-infiltrating APCs and higher CXCL17 expression in human epithelial ovarian cancer. Oncoimmunology 2019;8(12):e1665460.
[Crossref] [Google Scholar] [PubMed]
- Yuan L, Ye J, Fan D. The B7-H4 gene induces immune escape partly via upregulating the PD-1/Stat3 pathway in non-small cell lung cancer. Hum Immunol 2020;81(5):254-61.
[Crossref] [Google Scholar] [PubMed]
- Cheng C, Qu QX, Shen Y, Lv YT, Zhu YB, Zhang XG, et al. Overexpression of B7-H4 in tumor infiltrated dendritic cells. J Immunoassay Immunochem 2011;32(4):353-64.
[Crossref] [Google Scholar] [PubMed]
- Kryczek I, Wei S, Zou L, Zhu G, Mottram P, Xu H, et al. Cutting edge: Induction of B7-H4 on APCs through IL-10: Novel suppressive mode for regulatory T cells. J Immunol 2006;177(1):40-4.
[Crossref] [Google Scholar] [PubMed]
- Archer K, Ceradoy J, Coupet T, Bosiacki J, Song C, Weiss I, et al. Development and functional characterization of NC762, a novel therapeutic antibody targeting B7-H4, for the treatment of malignancies. Cancer Res 2021;81(13):3193.
- Fessler SP, Wang J, Collins SD, Qin L, Avocetien K, Xu L, et al. XMT-1660, a B7-H4-targeted Dolasynthen antibody-drug conjugate for the treatment of breast cancer. Cancer Res 2021;81:907.
- Chen C, Zhu WD, Xie F, Huang JA. Nuclear localization of B7-H4 in pulmonary adenocarcinomas presenting as a solitary pulmonary nodule. Oncotarget 2016;7(36):58563-8.
[Crossref] [Google Scholar] [PubMed]
- Song H, Xie W, Lian Q, Chen M, Xu R, Zeng S, et al. Inhibition of PI3K/AKT signaling pathway promotes the nuclear translocation of B7-H4. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2014;30(11):1121-4.
[Google Scholar] [PubMed]
- Sadun RE, Sachsman SM, Chen X, Christenson KW, Morris WZ, Hu P, et al. Immune signatures of murine and human cancers reveal unique mechanisms of tumor escape and new targets for cancer immunotherapy. Clin Cancer Res 2007;13(13):4016-25.
[Crossref] [Google Scholar] [PubMed]
- Yin Y, Shi L, Yang J, Wang H, Yang H, Wang Q. B7 family member H4 induces epithelial-mesenchymal transition and promotes the proliferation, migration and invasion of colorectal cancer cells. Bioengineered 2022;13(1):107-18.
[Crossref] [Google Scholar] [PubMed]
 aberation
aberation