- *Corresponding Author:
- Ting Wei
Department of Neurosurgery, The First Affiliated Hospital of Xi’an Jiaotong University, Xi’an 710061, China
E-mail: liangchen01@xjtu.edu.cn
| Date of Received | 26 February 2023 |
| Date of Revision | 13 February 2024 |
| Date of Acceptance | 04 July 2024 |
| Indian J Pharm Sci 2024;86(4):1239-1246 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Chronic subdural hematoma primarily affects the elderly, and its morbidity increases with the global rise in the elderly population. Atorvastatin, commonly prescribed for hypercholesterolemia and atherosclerosis, has shown therapeutic promise for chronic subdural hematoma, although its mechanism remains unclear. This study employs network pharmacology and reverse transcription-quantitative polymerase chain reaction to analyze and preliminarily verify atorvastatin's potential mechanism in treating chronic subdural hematoma. Analysis identified 19 potential targets of atorvastatin in chronic subdural hematoma treatment. Enrichment analysis revealed that these target genes predominantly regulate inflammation, angiogenesis, and coagulation/fibrinolysis. From the 19 target genes, matrix metalloproteinase 2, matrix metalloproteinase 9, interleukin-6, C-X-C motif chemokine ligand 8, and serpin family E member 1 were selected to verify atorvastatin's effect. Reverse transcription-quantitative polymerase chain reaction demonstrated that atorvastatin significantly suppressed the expression of these genes in human umbilical vein endothelial cells (p<0.01). These findings partially elucidate atorvastatin's potential mechanism in treating chronic subdural hematoma, offering research insights and a foundation for further study.
Keywords
Atorvastatin, chronic subdural hematoma, network pharmacology, inflammatory factor, hematoma membrane
Chronic subdural hematoma (CSDH) refers to an enclosed accumulation of blood and its breakdown products between the brain's surface and the dura mater. According to classical theory, CSDH arises from minor craniocerebral trauma, typically affecting the bridging veins connecting the brain to the draining dural venous sinuses[1]. Prevalent among the elderly population, CSDH ranks among the most prevalent neurosurgical conditions. Its incidence has steadily increased, possibly attributed to the expanding elderly demographic and increased use of antithrombotic medications[2], highlighting its growing significance in neurosurgery. The pathogenesis of CSDH is multifaceted, with inflammation being a pivotal component. After the initial formation of the hematoma, an inflammatory cascade is triggered, leading to recruitment of inflammatory cells, fibrinolysis, formation of the hematoma membrane, and release of angiogenic factors. These factors stimulate the growth of new blood vessels within the hematoma membrane. However, these newly formed vessels are fragile and highly permeable, promoting microhemorrhages and fluid exudation, ultimately resulting in fluid accumulation within the hematoma cavity over time[3]. Given the pivotal roles of inflammation and angiogenesis in CSDH pathogenesis, current drug therapies primarily target these processes using anti-inflammatory or anti-angiogenic agents. Atorvastatin, a statin commonly prescribed for hypercholesterolemia and atherosclerosis, has demonstrated pleiotropic effects, including antiangiogenic and anti-inflammatory properties[4,5], which are relevant to CSDH treatment. Initial clinical investigations suggest that atorvastatin may expedite CSDH absorption and improve neurological outcomes in affected patients[6-8]. Similar findings have been reported in animal studies[9,10]. However, the precise mechanism underlying atorvastatin's efficacy in CSDH treatment remains unclear. Network pharmacology is a method used to elucidate the multifaceted actions of drugs and their interactions with various targets[11]. It employs bioinformatic techniques to comprehensively map out a drug's molecular interactions, aiding in the identification of new drugs or new targets for existing drugs[12,13]. Due to these attributes, network pharmacology holds significant promise in uncovering the mechanisms of drug action. This study aims to employ network pharmacology to analyze the underlying mechanism of atorvastatin in CSDH treatment.
Materials and Methods
Selection of potential atorvastatin targets:
Potential targets of atorvastatin were sourced from the chEMBL (https://www.ebi.ac.uk/chembl/), NCBI PubChem Compound (https://www.ncbi.nlm.nih.gov/pccompound), and SwissTargetPrediction (http://www.swisstargetprediction.ch/) databases using 'atorvastatin' as the search term. All target genes were converted into standard gene names using the UniProt database (https://www.uniprot.org/), and duplicate genes from different databases were eliminated.
Selection of disease-related genes:
Disease-related genes for CSDH were retrieved from the GeneCards database (https://www.genecards.org/) using 'CSDH' as keywords, and from the DisGeNET database (https://www.disgenet.org/) using 'subdural hematoma' as keywords. Duplicate genes from different databases were excluded.
Prediction of potential targets of atorvastatin in the treatment of CSDH:
The potential target genes for atorvastatin and the related genes for CSDH were compared using venn diagrams generated by Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/), and the overlapping genes were identified as potential target genes for atorvastatin in CSDH treatment.
Pathway enrichment analysis:
R 4.2.1 software (R Studio, USA) was used for pathway enrichment analysis. The 'org.Hs.eg.db' package was utilised to obtain EntrezIDs for potential target genes. Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed for these potential target genes using the clusterProfiler, org.Hs.eg.db, enrichplot, and pathview packages, with a p-value cutoff of 0.05 and a q-value cutoff of 0.05. Results were visualised using the 'ggplot2' package. A network graph illustrating the relationship between target genes and pathways was created using Cytoscape 3.9.1 software (Cytoscape Consortium, United States of America (USA)).
Protein-Protein Interaction (PPI) analysis:
The Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) was employed for PPI analysis to reveal the interaction network among target genes at the translational level. The PPI network graph was constructed using Cytoscape 3.9.1 software (Cytoscape Consortium, USA).
Quantitative real-time Polymerase Chain Reaction (PCR) (Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR)) to detect the expression of target genes:
Based on previous studies and the PPI network, key genes involved in CSDH formation were selected from the target genes. RT-qPCR was performed to assess the expression of these genes in Endothelial Cells (ECs) before and after atorvastatin treatment. Human Umbilical Vein Endothelial Cells (HUVECs) were obtained from Procell Life Science & Technology Co., Ltd., (Wuhan, China, Cat No.CL-0122) and authenticated using Short Tandem Repeat (STR) profiling. HUVECs were cultured in F12K medium (Procell) supplemented with 10 % fetal bovine serum (Gibco) at 37° with 5 % CO2. Cells were seeded onto 6-well plates at a density of 3×105 cells/well. The atorvastatin treatment group was incubated in medium containing 0.1 µmol/l atorvastatin (Sigma-Aldrich, USA, Cat No. SML3030). Cells were harvested 24 h after drug treatment, lysed, and total Ribonucleic Acid (RNA) was isolated using an RNA fast 200 kit (Shanghai Fastagen Biotechnology Co., Ltd., China) according to the manufacturer's instructions. RNA was reverse-transcribed using a PrimeScript RT Master Mix kit (TaKaRa Bio, Inc. Japan). RT-qPCR was performed using SYBR Premix Ex Taq II (TaKaRa Bio, Inc.) on an iQ5 thermal cycler and analysed using iQ5 software, version 2.0 (Bio-Rad Laboratories, Inc., USA). Data were normalised to the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and alterations in gene expression were evaluated using the 2-??Ct method. The primers for these genes are shown in Table 1.
| Gene | Sequence |
|---|---|
| IL-6 | Forward, 5′- CTGGATTCAATGAGGAGAC -3′ |
| Reverse, 5′- ATTTGTGGTTGGGTCAGG -3′ | |
| MMP-2 | Forward, 5′- TATGACAGCTGCACCACTGA -3′ |
| Reverse, 5′- TCATCGTAGTTGGCTGTGGT -3′ | |
| CXCL-8 | Forward, 5′- AACACAGAAATTATTGTAAAG -3′ |
| Reverse, 5′ - CACTGATTCTTGGATACC -3′ | |
| MMP-9 | Forward, 5′- TGCCACTTCCCCTTCATCTT -3′ |
| Reverse, 5′- CGTCCTGGGTGTAGAGTCTC -3′ | |
| SERPINE-1 | Forward, 5′- GTGCTGGTGAATGCCCTCTAC -3 ′ |
| Reverse, 5′- CAGTGCTGCCGTCTGATTTGT -3′ | |
| GAPDH | Forward, 5′- TCAAGAAGGTGGTGAAGCAGG -3′ |
| Reverse, 5′ - TCAAAGGTGGAGGAGTGGGT -3′ |
Table 1: Sequences of primer for the quantitative polymerase chain reaction.
Statistical analysis:
Gene expression levels were expressed as mean ± standard deviation and analysed using SPSS 17.0 software (SPSS, Inc.). Student's t-test was employed for group comparisons, with p<0.05 considered statistically significant.
Results and Discussion
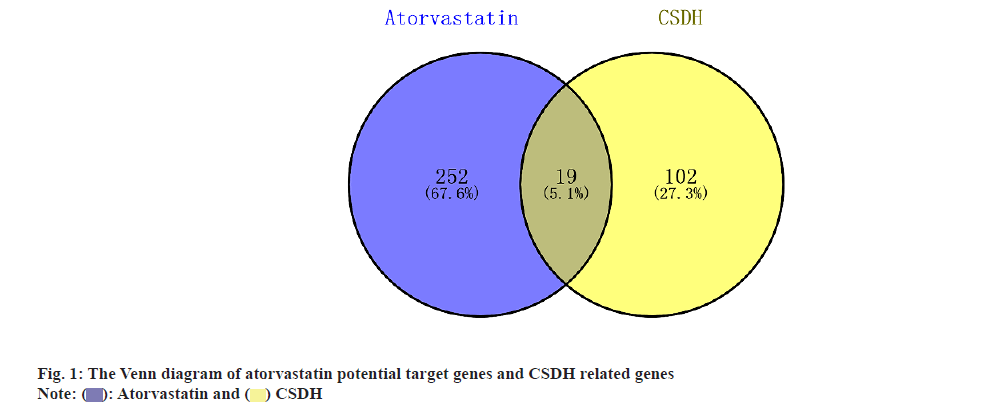
A total of 324 targets were initially identified from the chEMBL, National Center for Biotechnology Information (NCBI) PubChem compound, and SwissTargetPrediction databases, with 271 unique targets remaining after duplicate removal. From the GeneCards database, 109 CSDH-related genes were obtained, and an additional 3 genes were retrieved from the DisGeNET database. After eliminating duplicates, a total of 121 unique genes were identified. Following the intersection of atorvastatin potential target genes and CSDH-related genes, 19 potential targets of atorvastatin in the treatment of CSDH were identified, as shown in fig. 1 and Table 2.
| Gene symbol | Gene name |
|---|---|
| MMP-9 | Matrix metalloproteinase 9 |
| SERPINE-1 | Serpin family e member 1 |
| MMP-2 | Matrix metalloproteinase 2 |
| FGF-2 | Fibroblast growth factor 2 |
| CXCL-10 | C-X-C motif chemokine 10 |
| PLAT | Tissue-type plasminogen activator |
| AKT-1 | AKT serine/threonine kinase 1 |
| ANGPT-2 | Angiopoietin 2 |
| CCL-2 | C-C motif chemokine ligand 2 |
| CD-36 | CD36 molecule |
| CRP | C-reactive protein |
| CXCL-8 | C-X-C motif chemokine ligand 8 |
| EDN-1 | Endothelin 1 |
| F2 | Coagulation factor II |
| F5 | Coagulation factor V |
| IL-6 | Interleukin-6 |
| INS | Insulin |
| VWF | Von Willebrand factor |
| F7 | Coagulation factor VII |
Table 2: Potential targets of atorvastatin in the treatment of CSDH.
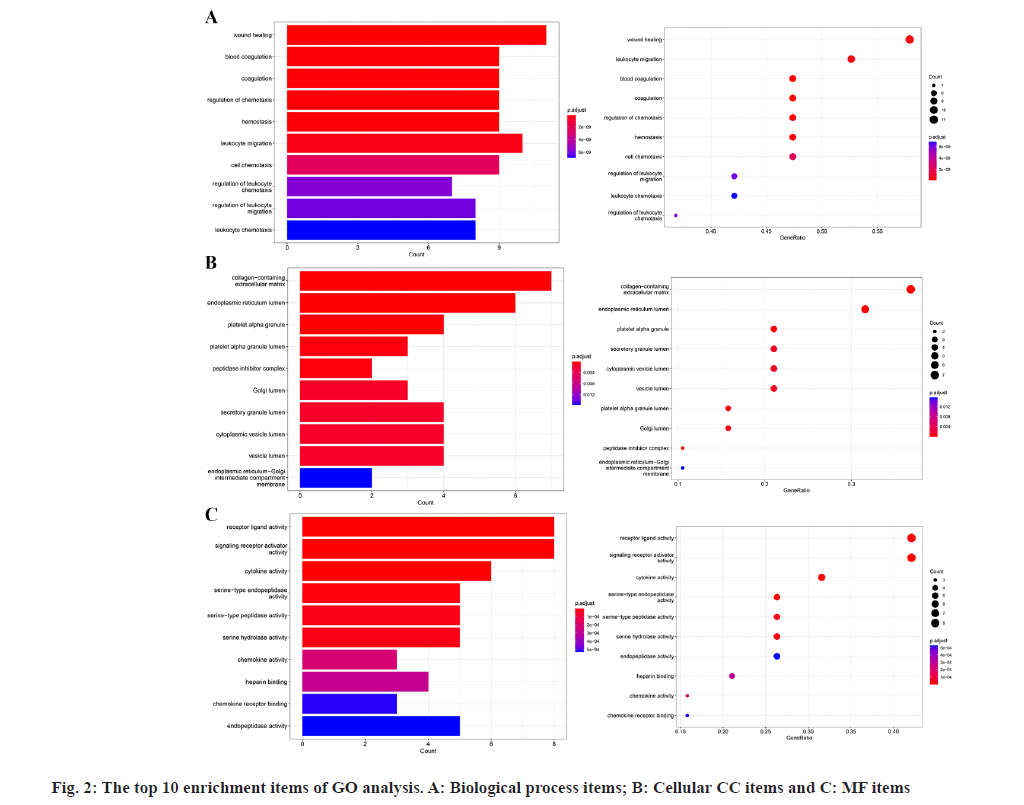
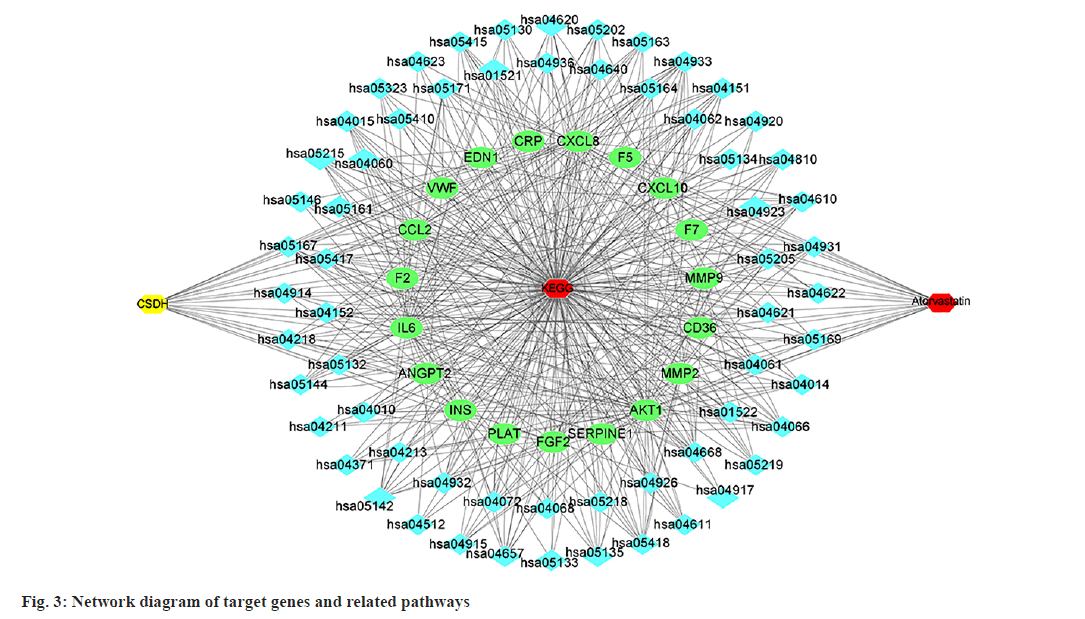
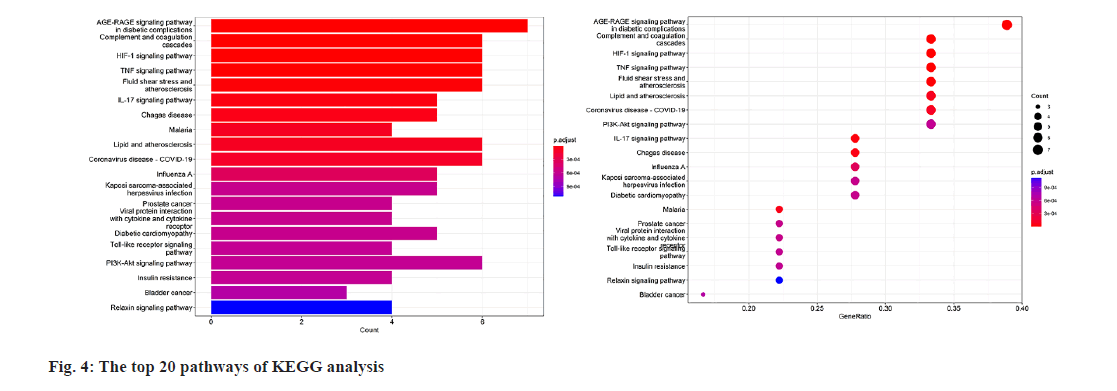
GO enrichment analysis of the 19 potential atorvastatin targets revealed 948 GO items, comprising 897 biological process items, 15 Cellular Component (CC) items, and 36 Molecular Function (MF) items. The top 10 enrichment items in each category are shown in fig. 2. The results suggest that the mechanism of atorvastatin in treating CSDH may involve processes such as wound healing, coagulation, chemotaxis, hemostasis, and leukocyte migration. MFs related to receptor ligand activity, cytokine activity, and serine hydrolase activity was also implicated. CCs such as the collagen-containing extracellular matrix, peptidase inhibitor complex, secretory granule lumen, and cytoplasmic vesicle lumen were involved. KEGG pathway enrichment analysis identified 62 pathways related to the potential targets of atorvastatin in CSDH treatment (fig. 3), with the top 20 pathways shown in fig. 4. These pathways are associated with the regulation of inflammatory responses, coagulation processes, and angiogenesis, including the Hypoxia-Inducible Factor 1 (HIF-1) signaling pathway, tumor necrosis factor (TNF) signaling pathway, complement and coagulation cascades, toll-like receptor signaling pathway, phosphatidylinositol 3 kinase (PI3K)- protein kinase B (Akt) signaling pathway, and the Interleukin-17 (IL-17) signaling pathway.
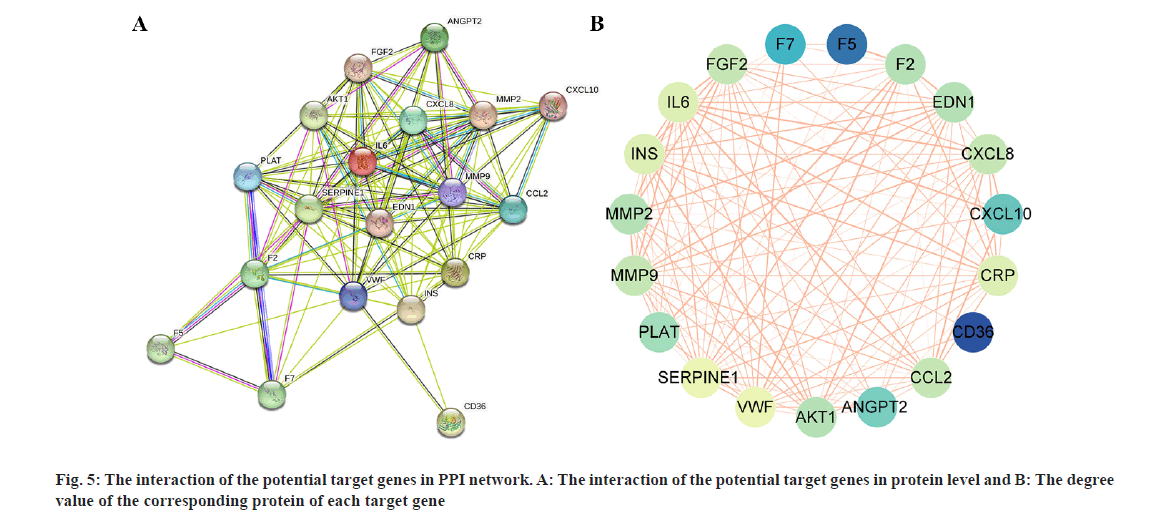
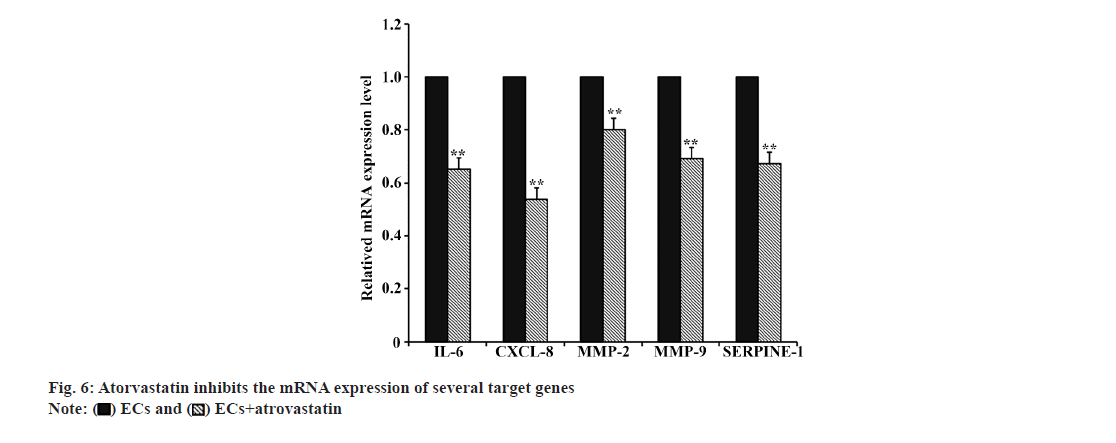
The PPI network (fig. 5A) illustrates the interaction of potential target genes at the protein level. Fig. 5B, displays the degree value of each target gene's corresponding protein, with lighter colors indicating higher degree values, suggesting greater importance in the network. Based on previous studies and the PPI network, Matrix Metalloproteinase (MMP)-2, MMP-9, IL-6, C-X-C motif chemokine ligand 8 (CXCL-8), and Serpin family E member 1 (SERPINE-1) were chosen from the 19 target genes to assess the impact of atorvastatin. As illustrated in fig.6, atorvastatin significantly suppressed the transcriptional expression of these genes in HUVECs (p<0.01).
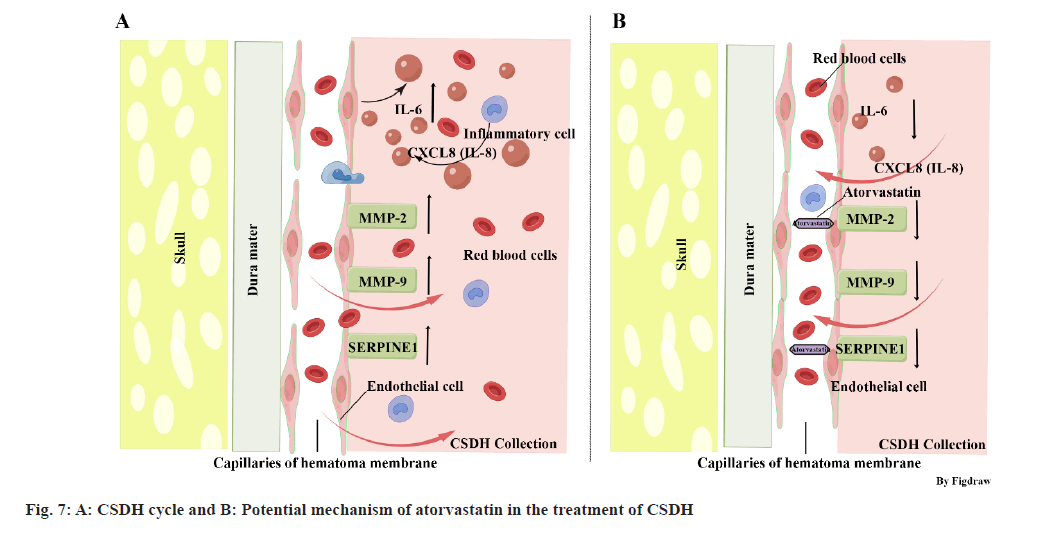
The pathophysiological understanding of CSDH has evolved over time. It is now widely accepted that initial bleeding in CSDH originates from bridging veins, which can be damaged due to factors such as trauma (especially in elderly individuals with brain atrophy), intracranial hypotension, or cranial surgery[1,14]. In response to this initial bleeding, inflammatory cells including neutrophils, lymphocytes, macrophages, and eosinophils are recruited to the subdural space to aid in tissue repair. Alongside tissue repair, these inflammatory cells also release inflammatory and angiogenic factors, promoting the formation of the hematoma membrane and new blood vessels within it. These new blood vessels are highly permeable, allowing blood components to exude into the hematoma cavity, leading to ongoing inflammatory activation and further development of the hematoma membrane (fig. 7A). This cycle of events is often referred to as the CSDH cycle[3]. In addition to inflammation and angiogenesis, dysregulated coagulation/fibrinolysis is also believed to play a role in CSDH development. Excessive fibrinolysis may contribute to continued hemorrhage and expansion of the CSDH[3], and markers of hyperfibrinolysis, such as tissue Plasminogen Activator (tPA), may serve as predictors for CSDH recurrence[15].
The GO enrichment analysis in this study revealed that the biological processes influenced by atorvastatin's target genes in CSDH treatment are closely linked to inflammation, angiogenesis, and coagulation/fibrinolysis. Among the top 10 enriched biological processes, wound healing is associated with angiogenesis and inflammation, while blood coagulation, coagulation, and hemostasis are linked to the regulation of coagulation and fibrinolysis. Additionally, regulation of chemotaxis, leukocyte migration, cell chemotaxis, regulation of leukocyte chemotaxis, regulation of leukocyte migration, and leukocyte chemotaxis are related to inflammation. These findings suggest that atorvastatin's mechanism in treating CSDH may involve the regulation of these three pathophysiological processes.
The PPI network analysis in this study revealed the interaction of potential target genes and their degree value. 5 genes (IL-6, CXCL-8, MMP-2, MMP-9, and SERPINE-1) were selected based on previous studies and the PPI network, as they play significant roles in the pathophysiological processes of inflammation, angiogenesis, and coagulation/fibrinolysis. IL-6 and CXCL-8, also known as IL-8, are crucial inflammatory agents secreted by various cell types, including fibroblasts, endothelial cells, and immune cells[14,16]. IL-6 is pivotal in the acute inflammatory response, immune cell differentiation, platelet production, and inflammatory cell recruitment[3]. CXCL-8 is a chemokine influencing immune cell migration and is considered a key cytokine in angiogenesis, promoting capillary tube formation, endothelial cell proliferation, and MMP-2 release[16]. Both IL-6 and CXCL-8 are found in high concentrations in CSDH fluid, particularly in recurrent cases[17,18]. MMPs are a family of proteolytic enzymes crucial in inflammation and angiogenesis. The imbalance of MMP-2 and MMP-9 expression is believed to relate to the instability of newly formed blood vessels, increasing the risk of bleeding[19]. MMP-2 and MMP-9 also participate in the inflammatory cascade by regulating various inflammatory mediators and their receptors[19]. Studies have shown significant elevation of MMP-2 and MMP-9 in CSDH fluid[20]. SERPINE-1, also known as Plasminogen Activator Inhibitor 1 (PAI-1), belongs to the serine protease inhibitor family, regulating fibrinolysis and inhibiting tPA and urokinase. SERPINE-1 is mainly produced by endothelial cells and participates in several pathophysiological processes, such as inflammation, oxidative stress, fibrosis, and macrophage adhesion/migration[21]. The results indicated that atorvastatin could inhibit the expression of IL-6, CXCL-8, MMP-2, MMP-9, and SERPINE-1 in HUVEC. Based on these findings, we hypothesize that atorvastatin may inhibit inflammatory reaction and angiogenesis, and regulate fibrinolysis by downregulating the expression of these genes, thereby reducing the formation of highly permeable blood vessels in the hematoma membrane, decreasing the infiltration of inflammatory cells, reducing endothelial cell damage, ultimately interrupting the CSDH cycle and promoting the liquefaction and absorption of hematoma (fig. 7B). However, considering the complexity of the CSDH formation mechanism and the regulatory effect of atorvastatin on other target genes, this hypothesis may only partially explain the mechanism of atorvastatin treatment of CSDH, requiring further confirmation through additional experiments.
According to the KEGG analysis results in this study, several classical pathways that regulate inflammation and angiogenesis, such as the HIF-1, TNF, IL-17, toll-like receptor, and PI3K-Akt signal pathways, may be involved in atorvastatin's regulation of target genes[22-26]. Additionally, pathways related to diabetes and atherosclerosis, which are known risk factors for CSDH, might also play a role in atorvastatin's treatment of CSDH[27,28]. This suggests a new research direction to elucidate atorvastatin's mechanism in CSDH treatment. Furthermore, infectious diseases and tumour-related pathways were also identified. Some studies have indicated that malignant tumours and infections are risk factors for CSDH recurrence[29,30]. The mechanisms of these diseases in CSDH development and atorvastatin's role in them require further research for confirmation.
However, there are limitations to this study. It is based on network pharmacology, which only provides a preliminary understanding of atorvastatin's mechanism in treating CSDH. Future research will need to further investigate atorvastatin's mechanism in treating CSDH both in vitro and in vivo. In conclusion, this study has identified the target genes and related signal pathways of atorvastatin in treating CSDH and has verified its effect on some target genes, which may provide a foundation for further research on the mechanism of atorvastatin in treating CSDH.
Conflict of interests:
The authors declared no conflict of interests.
References
- Nouri A, Gondar R, Schaller K, Meling T. Chronic Subdural Hematoma (cSDH): A review of the current state of the art. Brain Spine 2021;1:100300.
[Crossref] [Google Scholar] [PubMed]
- Rauhala M, Luoto TM, Huhtala H, Iverson GL, Niskakangas T, Öhman J, et al. The incidence of chronic subdural hematomas from 1990 to 2015 in a defined finnish population. J Neurosurg 2019;132(4):1147-57.
[Crossref] [Google Scholar] [PubMed]
- Edlmann E, Giorgi-Coll S, Whitfield PC, Carpenter KL, Hutchinson PJ. Pathophysiology of chronic subdural haematoma: Inflammation, angiogenesis and implications for pharmacotherapy. J Neuroinflammation 2017;14:1-3.
[Crossref] [Google Scholar] [PubMed]
- Araújo FA, Rocha MA, Mendes JB, Andrade SP. Atorvastatin inhibits inflammatory angiogenesis in mice through down regulation of VEGF, TNF-α and TGF-β1. Biomed Pharmacother 2010;64(1):29-34.
[Crossref] [Google Scholar] [PubMed]
- Bedi O, Dhawan V, Sharma PL, Kumar P. Pleiotropic effects of statins: New therapeutic targets in drug design. Naunyn Schmiedebergs Arch Pharmacol 2016;389:695-712.
[Crossref] [Google Scholar] [PubMed]
- Chan DY, Chan DT, Sun TF, Ng SC, Wong GK, Poon WS. The use of atorvastatin for chronic subdural haematoma: A retrospective cohort comparison study. Br J Neurosurg 2017;31(1):72-7.
[Crossref] [Google Scholar] [PubMed]
- Jiang R, Zhao S, Wang R, Feng H, Zhang J, Li X, et al. Safety and efficacy of atorvastatin for chronic subdural hematoma in Chinese patients: A randomized ClinicalTrial. JAMA Neurol 2018;75(11):1338-46.
[Crossref] [Google Scholar] [PubMed]
- He C, Xia P, Xu J, Chen L, Zhang Q. Evaluation of the efficacy of atorvastatin in the treatment for chronic subdural hematoma: A meta-analysis. Neurosurg Rev 2021;44:479-84.
[Crossref] [Google Scholar] [PubMed]
- Quan W, Zhang Z, Li P, Tian Q, Huang J, Qian Y, et al. Role of regulatory T cells in atorvastatin induced absorption of chronic subdural hematoma in rats. Aging Dis 2019;10(5):992.
[Crossref] [Google Scholar] [PubMed]
- Yang L, Li N, Yang L, Wang D, Qiang S, Zhao Z. Atorvastatin-induced absorption of chronic subdural hematoma is partially attributed to the polarization of macrophages. J Mol Neurosci 2022:1-9.
[Crossref] [Google Scholar] [PubMed]
- Zhou S, Bi J, Zhao W, Zhao J, Wan H, Wang S. Study on the mechanism of fentanyl in pain treatment based on network pharmacology. J Healthc Eng 2022;2022(1):4139200.
[Crossref] [Google Scholar] [PubMed]
- Zhang J, Jingjing ZH, Siqi HE, Jianxun WA. Efficacy of glucocorticoids, chloroquine and vitamin A on cytokine release syndrome: A network pharmacology study. J Tradit Chin Med 2022;42(1):116.
[Crossref] [Google Scholar] [PubMed]
- Zuhri UM, Purwaningsih EH, Fadilah F, Yuliana ND. Network pharmacology integrated molecular dynamics reveals the bioactive compounds and potential targets of Tinospora crispa Linn. as insulin sensitizer. Plos One 2022;17(6):e0251837.
[Crossref] [Google Scholar] [PubMed]
- Weigel R, Schilling L, Krauss JK. The pathophysiology of chronic subdural hematoma revisited: Emphasis on aging processes as key factor. GeroScience 2022;44(3):1353-71.
[Crossref] [Google Scholar] [PubMed]
- Katano H, Kamiya K, Mase M, Tanikawa M, Yamada K. Tissue plasminogen activator in chronic subdural hematomas as a predictor of recurrence. J Neurosurg 2006;104(1):79-84.
[Crossref] [Google Scholar] [PubMed]
- Li A, Varney ML, Valasek J, Godfrey M, Dave BJ, Singh RK. Autocrine role of interleukin-8 in induction of endothelial cell proliferation, survival, migration and MMP-2 production and angiogenesis. Angiogenesis 2005;8:63-71.
[Crossref] [Google Scholar] [PubMed]
- Hong HJ, Kim YJ, Yi HJ, Ko Y, Oh SJ, Kim JM. Role of angiogenic growth factors and inflammatory cytokine on recurrence of chronic subdural hematoma. Surg Neurol 2009;71(2):161-5.
[Crossref] [Google Scholar] [PubMed]
- Mustafa S, Koran S, AlOmair L. Insights into the role of matrix metalloproteinases in cancer and its various therapeutic aspects: A review. Front Mol Biosci 2022;9:896099.
[Crossref] [Google Scholar] [PubMed]
- Abdul-Muneer PM, Pfister BJ, Haorah J, Chandra N. Role of matrix metalloproteinases in the pathogenesis of traumatic brain injury. Mol Neurobiol 2016; 53(9): 6106-23.
[Crossref] [Google Scholar] [PubMed]
- Hua C, Zhao G, Feng Y, Yuan H, Song H, Bie L. Role of matrix metalloproteinase-2, matrix metalloproteinase-9, and vascular endothelial growth factor in the development of chronic subdural hematoma. J Neurotrauma 2016;33(1):65-70.
[Crossref] [Google Scholar] [PubMed]
- Kwak SY, Park S, Kim H, Lee SJ, Jang WS, Kim MJ, et al. Atorvastatin inhibits endothelial PAI-1-mediated monocyte migration and alleviates radiation-induced enteropathy. Int J Mol Sci 2021;22(4):1828.
[Crossref] [Google Scholar] [PubMed]
- Masoud GN, Li W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharmaceutica Sinica B. 2015;5(5):378-89.
[Crossref] [Google Scholar] [PubMed]
- Lebrec H, Ponce R, Preston BD, Iles J, Born TL, Hooper M. Tumor necrosis factor, tumor necrosis factor inhibition, and cancer risk. Curr Med Res Opin 2015;31(3):557-74.
[Crossref] [Google Scholar] [PubMed]
- Li X, Bechara R, Zhao J, McGeachy MJ, Gaffen SL. IL-17 receptor-based signaling and implications for disease. Nat Immunol 2019;20(12):1594-602.
[Crossref] [Google Scholar] [PubMed]
- Li TT, Ogino S, Qian ZR. Toll-like receptor signaling in colorectal cancer: Carcinogenesis to cancer therapy. World J Gastroenterol 2014;20(47):17699.
[Crossref] [Google Scholar] [PubMed]
- Isaji T, Osuka K, Ohmichi Y, Ohmichi M, Naito M, Nakano T, et al. Expression of angiopoietins and angiogenic signaling pathway molecules in chronic subdural hematomas. J Neurotrauma 2020;37(23):2493-8.
[Crossref] [Google Scholar] [PubMed]
- Erdogan B, Is M, Emon TS, Ceman D, Orakdogen M, Engin T. Retrospective analysis of 195 surgically treated cases of chronic subdural hematoma. Int J Clin Pract 2021;75(12):e15014.
[Crossref] [Google Scholar] [PubMed]
- Han SB, Choi SW, Song SH, Youm JY, Koh HS, Kim SH, et al. Prediction of chronic subdural hematoma in minor head trauma patients. Korean J Neurotrauma 2014;10(2):106.
[Crossref] [Google Scholar] [PubMed]
- Hori YS, Aoi M, Oda K, Fukuhara T. Presence of a malignant tumor as a novel predictive factor for repeated recurrences of chronic subdural hematoma. World Neurosurg 2017;105:714-9.
[Crossref] [Google Scholar] [PubMed]
- Dubinski D, Won SY, Trnovec S, Gounko K, Baumgarten P, Warnke P, et al. Recurrence of chronic subdural hematoma due to low-grade infection. Front Neurol 2022;13:1012255.
[Crossref] [Google Scholar] [PubMed]
 .
.
 .
.