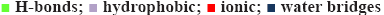- *Corresponding Author:
- A. Umamaheswari
Bioinformatics Centre, Department of Bioinformatics, SVIMS University, Tirupati-517 507, India
E-mail: amineni.maheswari@gmail.com
| Date of Submission | 01 September 2018 |
| Date of Revision | 23 November 2018 |
| Date of Acceptance | 18 March 2019 |
| Indian J Pharm Sci 2019;81(3):438-447 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In Mycobacterium tuberculosis, shikimate pathway is essential for amino acid biosynthesis, siderophores formation to overcome starvation, to survive in low oxygen conditions as well as for pathogen’s virulence and growth. 3-Dehydroquinate synthase of M. tuberculosis plays a vital role in the biosynthesis of aromatic amino acids and various secondary metabolites through shikimate pathway and is responsible for development of drug resistance. Thus, designing inhibitors towards this attractive drug target 3-dehydroquinate synthase to inhibit the synthesis of aromatic amino acids and essential secondary metabolites could prevent survival of this pathogen. In the present work, docking studies were performed using the 3-dehydroquinate synthase crystal structure against 1082 approved DrugBank compounds. The best DrugBank compound and substrate analogue (carbaphosphonate) were subjected to shape screening against 21 million compounds and resulted compounds constituted the AroB ligand-dataset. The library was subjected to rigid receptor docking, quantum polarized ligand docking, and induced fit docking followed by molecular mechanics-generalized born and surface area calculations resulted in two compounds possess the best scoring functions (XP GScore). Molecular dynamics simulations (50 ns) in the solvated model system determined consistency nature of AroB-lead 1 over the AroB-carbaphosphonate complex. Moreover, upon comparison of the proposed leads with the best DrugBank compound and carbaphosphonate, the leads showed favorable absorption, distribution, metabolism, excretion and toxicity properties within the range of 95 % FDA approved drugs, and showing better antagonist properties than the existing inhibitors. Hence these leads were proposed as novel inhibitors against tuberculosis.
Keywords
AroB, virtual screening, docking, molecular dynamics simulations and ADMET
Tuberculosis (TB), a major global problem caused by Mycobacterium tuberculosis, which is a life threatening intracellular pathogen in humans. It infects approximately one-third of the world’s population [1,2]. TB mostly affects people whose immunity is compromised. TB has many manifestations affecting bone, the central nervous system and many other organ systems, but it is primarily a pulmonary disease due to deposition of aerosol droplets contain M. tuberculosis onto lung alveolar surfaces [3]. Two types of drug resistant M. tuberculosis strains are currently recognized namely multi drug-resistant TB (MDR TB) and extensively drug-resistant TB (XDR TB) [4].
The recent emergence of multidrug-resistant strains of M. tuberculosis needs rapid development of new antimicrobial drugs to combat TB [5]. Proteins that are essential for the pathogen survival and absent in the host such as enzymes of the shikimate pathway are attractive targets for the development of new antiTB drugs [6,7]. Adverse effects of existing treatment options open up new challenges for researchers to discover novel lead molecules for treatment of TB [8].
In M. tuberculosis, the enzymes of the shikimate pathway are responsible for biosynthesis of three aromatic amino acids that include phenyl alanine, tyrosine, tryptophan and a range of other primary and secondary metabolites like folic acid, an essential cofactor for many enzymatic process and salicylate used for the biosynthesis of the siderophores and menaquinone, ubiquinone, naphthoquinones. The absence of this pathway in humans makes the enzymes of the pathway as the attractive drug targets [9-13].
AroB was identified as drug target based on comparative and subtractive proteomics approaches from 23 different strains of M. tuberculosis [14]. Dehydroquinate synthase (AroB) has long been regarded as a catalytic marvel because it is involved in the second step of the shikimate pathway. It catalyses the synthesis of dehydroquinate from 3-deoxy-d-arabino-heptulosonate 7-phosphate, a crucial step of this pathway resulted an essential cyclic compound, required for the synthesis of aromatic compounds [15]. As it is having role in aromatic amino acid biosynthesis, overcoming starvation and survival at hypoxic conditions. Hence, to design novel inhibitors to the emerging MDR and XDR TB, AroB was selected in the present work for inhibitor design.
Materials and Methods
Structure retrieval and binding site prediction:
Among the two available crystal structures, 3QBD and 3QBE of AroB of M. tuberculosis, the best resolute structure of 3QBE was considered in this study to propose antagonists through virtual screening, docking and molecular dynamics simulations. Crystal structure is available, but it’s not co-crystallized with substrate or inhibitor. Hence the orthologous structure of AroB of S. aureus co-crystallized with substrate analogue, carbaphosphonate (CBP) was selected to check the likelihood of the substrate binding site of AroB of M. tuberculosis using ClustalX v2.1.
Protein preparation:
Protein preparation tasks were performed with the protein preparation wizard. AroB crystal structure was imported to Maestro v11.1 (Schrödinger LLC, 2017) and prepared prior to docking in order to add hydrogen atoms, bond order and formal charge corrections, removed atomic clashes, adjustment of tautomerization and ionization states of protein. The hydrogen bonding network was optimized by reorienting the hydroxyl and thiol groups in the protein and perform other operations that are not part of the X-ray crystal structure refinement process. Finally the protein was subjected to energy minimization using Optimized Potential for Liquid Simulations (OPLS_3) force field with minimum RMSD for termination of minimization set was 0.3 Å and Grid box was generated to 10 Å×10 Å around active site residues of the AroB. Using the grid region placed on binding cleft of substrate, the unwanted water molecules were removed from the active site of target protein using Protein preparation wizard [16].
Ligand-preparation:
Inhibitors (published ligands) of AroB of M. tuberculosis were retrieved from literature and DrugBank v5.0 database, a unique bioinformatics and a drug chemoinformatics resource that combines detailed (chemical, pharmacological and pharmaceutical) data with comprehensive drug. The compounds were prepared to create 3-dimensional geometries, assign proper bond orders, and generate accessible tautomer and ionization states processed using LigPrep module [17]. Clustering was performed for the inhibitors based on fingerprints or properties by using Canvas v3.1 program. A representative member from each cluster was selected based on XPG score.
Glide (grid-based ligand docking with energetics) XP docking:
Docking is a procedure to predict the preferable binding orientation between the two molecules to form a stable complex. Docking calculations were carried out using Glide v7.4. The prepared and optimized ligands were flexibly docked in the grid box generated around active site residues of the protein using Monte Carlo-based simulated algorithm minimization method [18]. Glide Score (GScore) was used for representing binding affinity, binding orientation and ranking. Docking was implemented to retain lead molecules with better binding affinity and good binding orientation without stearic classes. 10 poses were generated during extra precision (XP) docking for each ligand and the best pose was retained after post docking minimization.
Exploring in-house library:
Based on the XP Gscore of the best docked DrugBank compound and substrate analogue were screened against an in-house library containing more than twenty-one million compounds from eMolecules, ChemBank, ChemPDB, KEGG ligand, Unannotated NCI, AntiHIV NCI, Drug likeness NCI, AkosGmbh, Asinex and TimTec databases [19]. The best docked two compounds as the structural anologues for DrugBank compound and substrates were obtained from the inhouse library. These best docked compounds and CBP (crystal substrate) were imported as AroB liganddataset to Maestro v11.1 for virtual screening workflow to dock with AroB of M. tuberculosis using Glide with defined pH range 7.0±2.0 by applying QikProp v5.1, Lipinski’s filter and reactive filter.
Docking studies:
AroB inhibitor dataset was docked with active site residues of AroB [20]. Virtual screening is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures, which are most likely to bind to a drug target. Schrödinger virtual screening workflow uses three flexible docking methods, namely Glide high throughput virtual screening (HTVS), standard precision (SP) and XP docking [21]. The top ligand molecules obtained through XP docking were compared with literature and existing drug bank compounds and CBP (substrate analogue) to propose AroB inhibitors.
The top lead molecules obtained through XP docking were re-docked using quantum polarized ligand docking (QPLD). Prime/MM-GBSA calculation was performed to compute the free energy of ligand binding (ΔG) for the top lead molecules with the common target by accessing both XP and QPLD complexes using Prime v3.2. The similar docking protocols followed by MM-GBSA calculations would be performed for selected inhibitors. The lead molecules with better XPG values were compared with existing ligands and proposed as potential inhibitors of their corresponding drug target. Binding free energies and binding modes of the proposed lead molecules would be reassessed using induced fit docking (IFD), which implements both receptor and ligand flexibility. The combination of molecular docking and Prime/MM-GBSA simulation can not only be used to rapidly and accurately predict the binding-free energy and rank the ligands but also provide a novel strategy for lead discovery and optimization towards the target.
Molecular dynamics:
Molecular dynamics simulations were performed for 50 ns to analyse the conformational stability of AroBCBP and-AroB-lead 1 complex in the solvated model system embedded with ordered water molecules using Desmond v4.8. Inter-molecular and intra-molecular interactions that influence the stability of protein-ligand complex were analysed using molecular dynamics. The solvated system was neutralized with counter ions and physiological salt concentration was limited to 0.15 M. The protein-ligand complex system was assigned with optimized potentials for liquid simulations-AA (OPLS-AA) 2005 force field [22]. The system was specified in periodic boundary conditions, the particle mesh Ewald method was applied for electrostatics. Lennard-Jones interactions cutoff was set to 10 Å and SHAKE algorithm was employed for limiting movement of all covalent bonds involving hydrogen atoms. The energy minimization of the solvated system was passed through a six-step relaxation protocol prior to molecular dynamics simulations [23]. In the earlier stage, only solvent molecules were allowed to minimize, while the protein and lead 1 are kept fixed. Then the entire system was minimized in the later stages using a hybrid method of steepest descent and limited-memory Broyden-Fletcher-Goldfarb-Shanno algorithm [24]. For restraining the non-hydrogen solute atoms, at a temperature of 10 K with a thermostat relaxation constant of 0.1 ps a short period of 12 ps simulation was carried out in the NVT ensemble. Another short period of 12 ps simulation in NPT ensemble using 10 K temperature (with thermostat relaxation constant of 0.1 ps and barostat relaxation constant of 50 ps) for restraining the non-hydrogen solute atoms. Then in the NPT ensemble solute nonhydrogen atoms were restrained for 24 ps simulations at a temperature of 300 K (with a thermostat relaxation constant of 0.1 ps; barostat relaxation constant of 50 ps) and 24 ps simulations in the NPT ensemble were carried out with no restrains at 300 K temperature (with a thermostat relaxation constant of 0.1 ps; barostat relaxation constant of 2.0 ps). Berendsen thermostats and barostats were used to control the temperatures and pressures during the initial simulations [25].
Following the relaxation of solvated system, a 50 ns of dynamics production run in the NPT ensemble (at temperature of 300 K, with thermostat relaxation time of 1.0 ps; 1.01 bar pressure, with barostat relaxation time of 2.0 ps) using a Nose-Hoover thermostat and Martyna-Tobias-Klein barostat was performed. Energy and trajectory of the atomic coordinates data were recorded with a time interval of 4.8 ps. The protein- CBP dock complexes were ruled out by energy potential, RMSD, RMSF, inter-molecular hydrogen bond interactions monitoring.
Results and Discussion
The 3-dehydroquinate synthase (AroB) is a Zn2+- dependent metalloprotein of M. tuberculosis involved in aromatic amino acid biosynthesis and secondary metabolite production to overcome starvation and survival at hypoxic conditions inside the human macrophages [26]. AroB protein of S. aureus having the substrate analogue CBP (1XAI-CBP) was selected for the active site prediction. CBP binds to C-terminal domain of AroB leading to domain closure [27]. AroB protein of M. tuberculosis is having 76 % query coverage and 35 % identity with S. aureus. Active site residues shared 100 % identity in both the sequences and were represented in red colour boxes (figure 1). Hence, Asp-138, Lys-144, Asn-154, Lys-189, Lys-228, Arg-242, His-249 and Lys-323 were considered as an active site residues of AroB of M. tuberculosis.
The energies of prepared crystal structure of AroB (3QBE) of M. tuberculosis were minimized by protein preparation wizard. Active site residues were defined around grid generated within the AroB protein. DrugBank compounds for AroB is more in number so that clustering was performed for structural analogues, resulted 11 clusters were formed. From each cluster centroid molecule were chosen to perform docking with substrate analogue (CBP). About 1082 prepared DrugBank compounds and one substrate analogue (CBP) were docked with AroB active site residues. Among the 1082 dock complexes, pravastatin possess the least XP GScore of –13.074 kcal/mol and CBP possesses XP Gscore of –10.665 kcal/mol with good bonded and non-bonded interactions with inhibitor binding site residues shown in Table 1 (figure 2). The best docked pravastatin and CBP were chosen for screening against in-house library database. Six thousand structure analogues obtained from the in-house library, pravastatin, substrate analogue (CBP) a total of 6002 compounds of AroB were imported to form an inhibitor dataset for docking.
| Existing inhibitors and leads | XP GScore (kcal/mol) |
|---|---|
| Carbaphosphonate | –10.665 |
| Provastatin | –13.074 |
| Lead 1 | –13.889 |
| Lead 2 | –13.107 |
Table 1: Molecular Docking Score of Carbaphosphonate, Pravastatin and Leads
Rigid receptor docking, QPLD and IFD protocols were employed to predicted the scoring and binding interactions between AroB and the ligands. In virtual screening a large number of molecules are ranked according to their likelihood to be bioactive compounds, with the aim to enrich the top fraction of the resulting list [25]. Out of 6002 ligands docked in HTVS method, top ranked 600 ligands were re-docked using SP method. Similarly top ranked 60 ligands were obtained through SP method were re-docked using XP docking method, 30 docked complexes were obtained with docking scores. Among them, two leads possessed better scoring function than the pravastatin and CBP (substrate analogue; figure 2). The lead complexes were re-scored with Prime/MM-GBSA. The re-scoring was performed as it is proved by various research groups that Prime/MM-GBSA re-scoring of docking complex (ΔG) showed better correlation to their experimental binding affinity compared to XP Gscore [26].
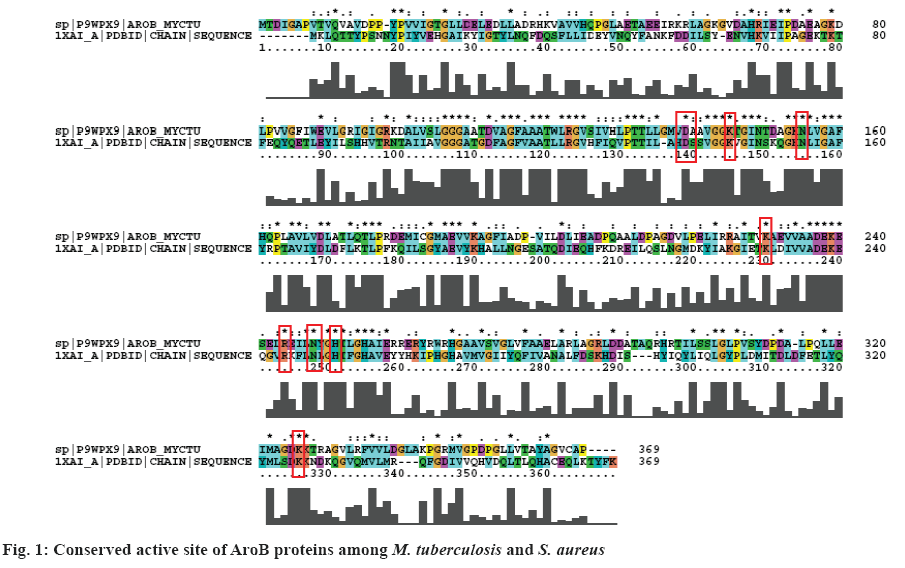
The docking strategy resulted CBP formed eight hydrogen bonds with active site residues such as Asp- 138, Lys-144(2), Lys-228, Arg-242, His-249(2), and Lys-323. Seven residues such as Asp-138, Lys-144, Asn-154, Lys-228, Arg-242, His-249, and Lys-323 showed non-bonded interactions.
Lead 1 (OC[C@@H](C)[C@H]1CC[C@H]([C@@] 12C)[C@@H](O)CCC2) formed six hydrogen bonds, one salt bridge, one metal co-ordination and fifteen van der Waal’s interactions with active site residues of AroB. Lys-78, Asp-138, Lys-144, Arg-242 (2) and Asn-246 are involved in hydrogen bond interactions; salt bridge with Lys-228; and one metal co-ordination with Zn; Glu-70, Gly-107, Ala-108, Asp-111, Leu-134, Ala-139, Asn-148, Lys-153, Asn-154, Lys-189, Leu- 245, His-249, His-253, Glu-256 and His-265 were involved in van der Waal’s interactions.
Lead 2 (CC1(C)[C@@H](C2)Cc(c3[C@H]12)noc3) formed six hydrogen bonds with active site residues Asp-138, Lys-144, Asn-154, Arg-242 (2) and Asn-246; two salt bridge interactions with Lys-144 and Lys-228; one pi-cation interaction with Lys-78 and one metal coordination with Zn. Fifteen residues Glu-75, Asp-111, Leu-134, Ala-139, Thr-145, Gly-146, Lys-143, Leu- 245, His-249, His-253, Glu-256, Tyr-261, His-265, Lys-323 and Lys-324 were involved in van der Waal’s interactions.
The proposed two leads and CBP were surrounded by inhibitor binding site residues of AroB of M. tuberculosis. Among the proposed two leads, lead 1 shared similar interactions with AroB of M. tuberculosis as the DrugBank compound and substrate analogue. Both the leads also deciphered two hydrogen bonds with the key catalytic residue Arg- 242 and metal co-ordination with Zn, the same was observed with the pravastatin and CBP.
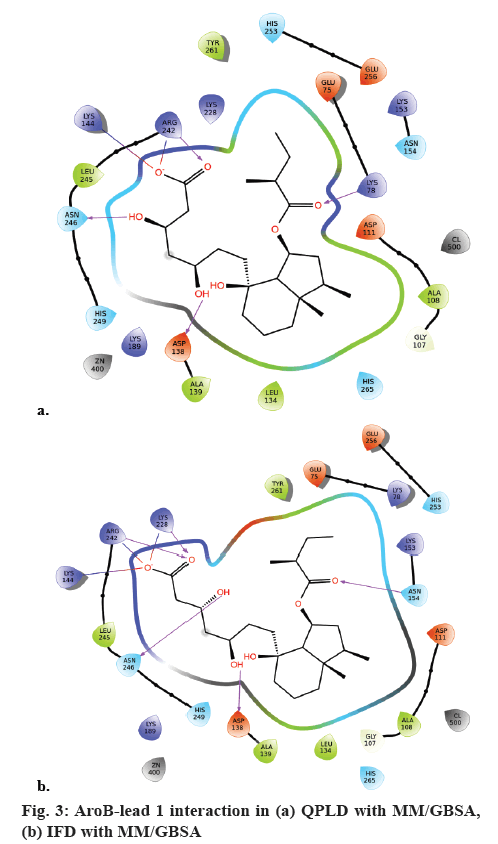
QPLD was employed to compute the atomic partial charges of the leads through quantum mechanical and molecular mechanical (QM/MM) calculations. The two leads, substrate analogue and pravastatin were subjected to re-dock with AroB using QPLD for evaluating relative binding interactions and strength of each potential lead with AroB after accurate charge calculation through hybrid quantum mechanics and molecular mechanics method. The compounds were ranked based on QPLD XP Gscore. Two leads showed the highest binding affinity towards AroB than the existing substrate analogue and pravastatin of AroB. Among them lead-1 showed the highest binding affinity towards AroB with QPLD XP Gscore of -14.036 kcal/ mol (figure 3a). Active site residues and their flexibility of AroB were considered for IFD protocol in Schrödinger. Lead molecule interacts with binding site residues of AroB by undergoing side chain or backbone conformational change or both. These conformational changes allow the AroB to generate closest conformer to the shape and binding mode of the leads. AroB lead interaction energies and total energy of the system was calculated as IFD score. Based on the IFD scores the poses generated were ranked. Similar binding interactions were observed in all the dock complexes with the leads docked around the AroB binding site residues. IFD had more interactions and scores than XP and QPLD (XP Gscore as –10.81 kcal/mol). AroB showed five hydrogen bonds with lead 1 in the IFD docked complex (figure 3b).
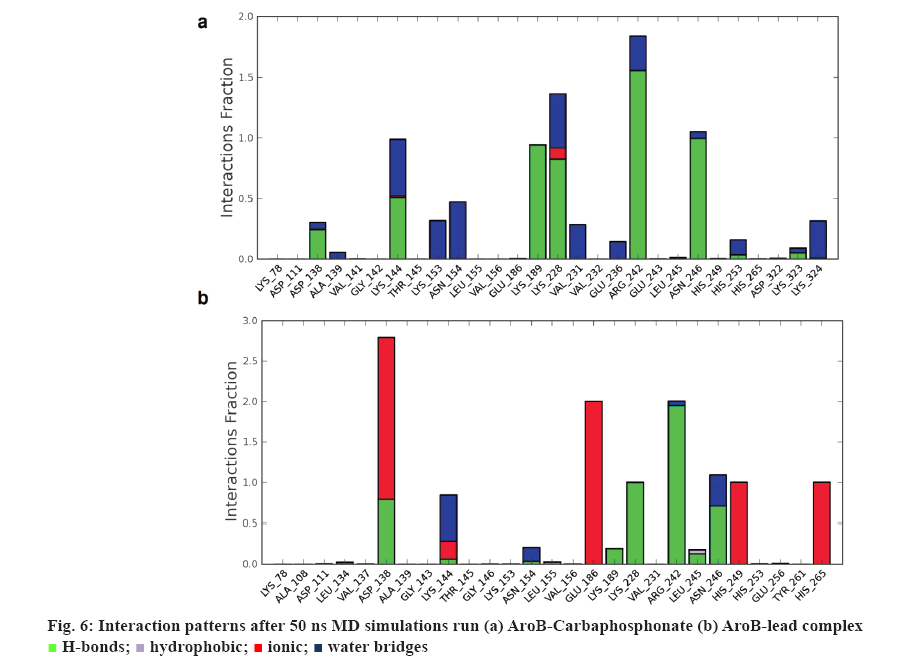
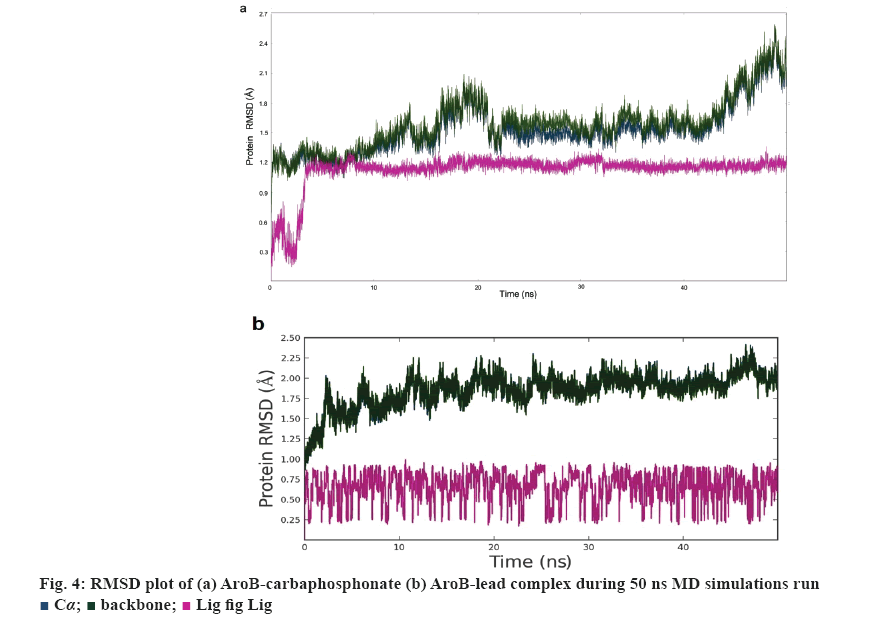
AroB-CBP complex has the average total energy of –103268.55 kcal/mol and AroB-lead complex has an average total energy of –103305.95 kcal/ mol. Average RMSD for AroB backbone and CBP were 1.58 Å and 1.12 Å and AroB backbone and lead were 1.85 Å and 0.66 Å, respectively which is stable throughout all 10416 trajectories (figures 4a-b). Average RMSF for backbone and side chain of AroB in accommodating CBP were 0.91 Å and 1.36 Å AroB in accommodating lead were 0.93 Å and 1.40, respectively (figure 5). Both lead and CBP exhibited hydrogen bonds with Asp- 138, Lys-144, Lys-189, Lys-228, Lys-242, Lys-323 (figure 6). CBP shows ionic bond with Lys-228, six water mediated hydrogen bonds with Asp-138, Lys-144, Lys- 189, Lys-228, Lys-242, Lys-323 with the key binding site residues in forming a stable complex (figure 6a). Lead exhibited three ionic bonds with Asp-138, Lys-144 and His-249, three water mediated hydrogen bonds with Lys-144, Asp-154, Arg-242 with the key binding site residues in forming a stable complex (figure 6b).
Average RMSD and RMSF of lead 1 and CBP having stable complex were within 2.5 Å of 50 ns MD simulations. Among them, lead 1 exhibited the least XP Gscore and total energy compared with substrate analogue CBP. AroB-lead 1 shows more contacts with active site residues than the AroB- CBP complex.
In summary, two leads were finally obtained as the outcomes the study with better binding affinity in terms of XPG scores, good structural properties with molecular contacts, pharmacological properties than the existing inhibitors and substrate. These absorption, distribution, metabolism, and excretion/toxicity properties of the proposed leads fall within the range of 95 % of FDA approved drugs were presented in Table 2 and 3. Aromatic amino acid biosynthesis is essential for the bacterium to survival, especially inside the CD4 cells by endogenous tryptophan synthesis. The ability of bacterial cells to replicate in the macrophages at hypoxic conditions is promoted by the biosynthesis and also lysis of phago-lysosome complex in macrophages. It also influences production of the secondary metabolites such as, siderophores, menaquinones, ubiquinone, napthoquinones, paraamino benzoic acid, which are essential for the virulence nature of the pathogen. Siderophores of bacterium are important for iron uptake from the host and essential for replication of M. tuberculosis in human macrophages. The study states that the proposed 2 leads showed the best binding orientation, binding affinity with strong hydrogen bond network, salt bridge interactions, good van der Waal’s interactions with active site residues (Asp-138, Lys-144, Lys-154, Lys-189, Lys-144, Lys- 228, Arg-242, His-249) of AroB-lead than the AroBCBP. Lead possess least potential energy (-129939.20 kcal/mol) when compared with CBP. The AroB-lead complex has an average RMSD and RMSF of the leads were within 2.5 Å of 50 ns MD to block the functional activity by interacting with the inhibitor binding site residues of AroB than the AroB- CBP complex. Hence, these leads serve as novel therapeutics if synthesized and validated in animal models.
| Compounds | M.W. | Rotor | SASA | FOSA | WPSA | PISA | Vol | Donor HB | Accept HB | IP | EA | Glob |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbaphosphonate | 270.17 | 8 | 440.7 | 108 | 3.54 | 0 | 748.15 | 6 | 11.15 | 10.4 | -0.33 | 0.9042 |
| Pravastatin | 424.53 | 13 | 690.6 | 373 | 0 | 91.00 | 1318.646 | 3 | 8.1 | 9.11 | -0.03 | 0.8420 |
| Lead 1 | 428.56 | 13 | 704.8 | 500 | 0 | 0 | 1355.331 | 3 | 7.15 | 10.5 | -1.07 | 0.8402 |
| Lead 2 | 423.52 | 13 | 696.0 | 411 | 0 | 44.01 | 1324.4 | 2 | 7.9 | 9.78 | 0.42 | 0.8379 |
| Parameters | Range 95 % of drugs | |||||||||||
| MW=molecular weight | (130.0/725.0) | |||||||||||
| Rotor=no. of rotation bonds | (0.0/15.0) | |||||||||||
| SASA=total solvent accessible surface area | (300.0/1000.0) | |||||||||||
| FOSA=hydrophobic solvent accessible surface area | (0.0/750.0) | |||||||||||
| PISA=carbon Pi solvent accessible surface area | (0.0/450.0) | |||||||||||
| Volume=molecular volume (A^3) | (500.0/2000) | |||||||||||
| Donor=donor -hydrogen bonds | (0.0/6.0) | |||||||||||
| AccptHB=acceptor-hydrogen bonds | (2.0/20.0) | |||||||||||
| IP (eV)=ionization potential | (7.9/10.5) | |||||||||||
| EA (eV)=electron affinity | (-0.9/1.7) | |||||||||||
| Glob=globularity | (0.75/0.95) | |||||||||||
Table 2: Pharmacological Descriptors of Carbaphosphonate, Pravastatin and Leads
| Compounds | Log P o/w | Log S | CI Log S | Log BB | RuleOf 5 | Rule of 3 | LogKp | LogKhSa |
|---|---|---|---|---|---|---|---|---|
| Carbaphosphonate | -1.02 | -1.229 | -0.854 | -2.592 | 1 | 1 | -6.814 | -1.436 |
| Pravastatin | 3.009 | -3.773 | -4.735 | -2.351 | 0 | 2 | -4.127 | -0.107 |
| Lead 1 | 3.639 | -4.266 | -4.735 | -2.146 | 0 | 0 | -4.031 | -0.134 |
| Lead 2 | 3.1 | -4.188 | -3.956 | -2.509 | 0 | 1 | -4.558 | -0.037 |
| Parameters | Range 95% of Drugs | |||||||
| Log P o/w = log P for octavo/water | (1.8/6.5) | |||||||
| Log S = log S for aqueous solubility | (6.5/0.5)) | |||||||
| CI Log S= log S - conformation independent | (6.5/0.5) | |||||||
| Log BB= log BB for brain/blood | (-3.0/1.2) | |||||||
| Log KP = log KP for skin permeability | (KP in cm/hr) | |||||||
| Log KhSa= log Khsa Serum Protein Binding | (-2.5/1.5) | |||||||
| Lipinski’s Rule of 5 Violations | (maximum is 4) | |||||||
| Jorgensen Rule of 3 Violations | (maximum is 3) | |||||||
Table 3: Predicted ADME/T Properties of Carbaphosphonate, Pravastatin and Leads
Acknowledgements
The authors are grateful to DBT, Ministry of Sciences and Technology, Govt. of India for providing studentship and traineeship facility to carry out the research work at BIF (BT/BI/25/037/2012BIFSVIMST).
References
- de Mendonça JD, Ely F, Palma MS, Frazzon J, Basso LA, Santos DS. Functional Characterization by Genetic Complementation of aroB-Encoded Dehydroquinate Synthase from Mycobacterium tuberculosis H37Rv and its Heterologous Expression and Purification. J Bacteriol 2007;189(17):6246-52.
- Smith I. Mycobacterium tuberculosis Pathogenesis and Molecular Determinants of Virulence. Clin Microbiol Rev 2003;16(3):463-96.
- Zumla A, Nahid P, Cole ST. Advances in the development of new tuberculosis drugs and treatment regimens. Nat Rev Drug Discov 2013;12(5):388-404.
- Müller B, Borrell S, Rose G, Gagneux S. The Heterogeneous Evolution of Multidrug Resistant Mycobacterium tuberculosis. Trends Genet 2013;29(3):160-9.
- Nazir T, Abraham S, Islam A. Emergence of Potential Superbug Mycobacterium tuberculosis, Lessons from New Delhi Mutant-1 Bacterial Strains. Int J Health Sci (Qassim) 2012; 6(1):87-94.
- de Mendonça JD, Adachi O, Rosado LA, Ducati RG, Santos DS, Basso LA. Kinetic mechanism determination and analysis of metal requirement of dehydroquinate synthase from Mycobacterium tuberculosis H37 Rv: An essential step in the function based rational design of anti TB-drugs. Mol Biosyst 2011;7(1):119-28.
- Ducati RG, Basso LA, Santo DS. Mycobacterial shikimate pathway Enzymes as Targets for Drug Design. Curr Drug Targets 2007;8(3);423-35.
- Goyal T, Anishetty S. Anin silico study on molecular level interactions of host Siderocalin with siderophores from Mycobacterium tuberculosis and other bacterial species. BMC Infect Dis 2014;14(Suppl 3):O15.
- Reddy PV, Puri RV, Chauhan P, Kar R, Rohilla A, Khera A, et al. Disruption of Mycobactine Biosynthesis Leads to Attenuation of Mycobacterium tuberculosis for Growth and Virulence. J Infect Dis 2013;15;208(8):1255-65.
- Jones CM, Wells RM, Madduri AV, Renfrow MB, Ratledge C, Moody DB, et al. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proc Natl Acad Sci USA 2014;111(5):1945-50.
- Gillespie SH. Evolution of Drug Resistance in Mycobacterium tuberculosis: Clinical and Molecular Perspective. Antimicrob Agents Chemother 2002;46:267-74.
- Dairi T. An alternative menaquinone biosynthetic pathway operating in microorganisms: an attractive target for drug discovery to pathogenic Helicobacter and Chlamydia strains. J Antibiot 2009;62(3):47-52.
- Moura-Alves P, Faé K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature 2014;512:387-92.
- Pasala C, Chilamakuri CSR, Katari SK, Nalamolu RM, Bitla AR, Umamaheswari A. An in silico study: Novel targets for potential drug and vaccine design against drug resistant H. pylori. Microb Pathog 2018; 122:156-61.
- Jorgensen WL, Maxwell DS, Tirado Rives J. Development and Testing of the OPLS All Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J Am Chem Soc 1996;118:11225-36.
- Pradeep N, Munikumar M, Swargam S, Hema K, Sudheer Kumar K. Combinations of e-pharmacophoremodeling, multiple docking strategies and molecular dynamic simulations to discover of novel antagonists of BACE1. J Biomol Stuct Dyn 2015;1:129-33.
- Umamaheswari A, Kumar MM, Pradhan D, Marisetty H. Docking studies towards exploring antiviral compounds against envelope protein of yellow fever virus. Interdiscip Sci 2011;3(1):64-77.
- Sandeep S, Priyadarshini V, Pradhan D, Munikumar M, Umamaheswari A. Docking and molecular dynamics simulations studies of human protein kinase catalytic subunit alpha with antagonist. J Clin Sci Res 2012;1:15-23.
- Hema K, Sandeep S, Pradeep N, Umamaheswari A. In silico agonist for human extracellular superoxide dismutase SOD3.Online J Bioinform 2016;17(1):29-40.
- Brooks WH, Danieel KG, Sung SS, Guida WC, Computational validation of the importance of absolute stereochemistry in virtual screening. J Chem Inf Model 2008;48:639-45.
- Madhulatha NR, Pradeep N, Sandeep S, Hema K, Chiranjeevi P, Kumar KS, et al. E-Pharmacophore Model Assisted Discovery of Novel Antagonists of nNOS. Biochem Anal Biochem 2017;6:1.
- Katari SK, Natarajan P, Swargam S, Kanipakam H, Pasala C, Umamaheswari A. Inhibitor design against JNK1 through e-pharmacophore modeling docking and molecular dynamics simulations. J Recept Signal Transduct Res 2016;36(6):558-71.
- Chiranjeevi P, Swargam S, Pradeep N, Hema K, Kumar SK, Madhulatha NR, et al. Inhibitor design for VacA toxin of Helicobacter pylori. J Proteomics Bioinform 2016;9:220-25.
- Webby CJ, Jiao W, Hutton RD, Blackmore NJ, Baker HM, Baker EN, et al. Synergistic Allostery, a Sophisticated Regulatory Network for the Control of Aromatic Amino Acid Biosynthesis in Mycobacterium tuberculosis. J Biol Chem 2010;285(40):30567-76.
- Natarajan P, Swargam S, Hema K, Vengamma B, Umamaheswari A. E-Pharmacophore Based Virtual Screening to Identify Agonist for PKA-Cα. Biochem Anal Biochem 2015;4:222.
- Pradhan D, Priyadarshini V, Munikumar M, Swargam S, Umamaheswari A, Bitla A. Para-(benzoyl)-Phenylalanine as a Potential Inhibitor against LpxC of Leptospira Spp.: Homology Modeling, Docking and Molecular Dynamics Study. J Biomol Struct Dyn 2014;32:171-85.
- Nichols CE, Ren J, Leslie K, Dhaliwal B, Lockyer M, Charles I, et al. Comparison of Ligand-induced Conformational Changes and Domain Closure Mechanisms, Between Prokaryotic and Eukaryotic Dehydroquinate Synthases. J Mol Biol 2004;343;533-46.
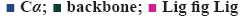
 the green vertical bars in both RMSF plots give an insight of residual interactions throughout the 50 ns dynamics simulations
the green vertical bars in both RMSF plots give an insight of residual interactions throughout the 50 ns dynamics simulations